A new study published in Nature reports how researchers verified the intricate process of disaggregating entangled proteins by analyzing chaperone enzyme ClpB at a single-molecule level. The C-Trap® correlated optical tweezers and confocal microscopy enabled them to analyze how the member of the Hsp100 chaperone family translocates looped polypeptide substrates in a processive manner.
The findings serve to explain how disaggregating proteins disentangle and refold polypeptides within protein aggregates – structures that are found in several diseases. Congratulations to Prof. Dr. Sander Tans at AMOLF, his lab, and all the authors involved in this work!
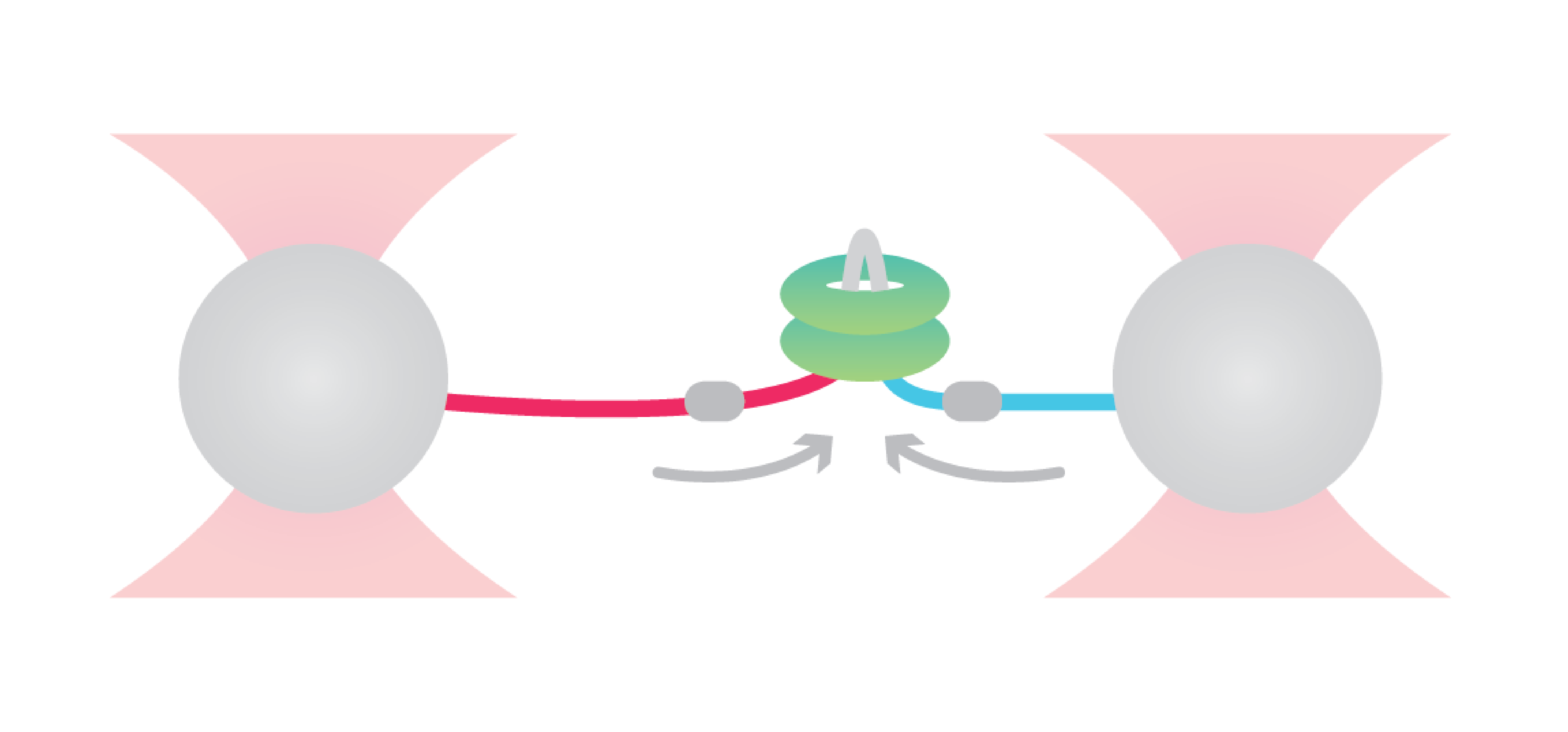
To uncover the enzymatic activity of ClpB, Avellaneda et al. considered different hypothetical translocation models as a starting point, including looped dual-strand insertion models.
The researchers first tethered the bacterial maltose-binding protein (MBP) between two optically trapped beads through DNA handles. Next, they introduced a fluorescently labeled ClpB and assessed distance-changes between the two MBP-termini upon translocation.
Observing first that ClpB applied two-arm polypeptide translocation, they confirmed a dual-strand insertion model. The approach further revealed that ClpB alternates between single-strand translocations and two-fold faster dual-strand translocations with occasional back slips of the single-arms. On top of that, they found that the transition between these two modes depended on substrate refolding that ultimately blocked the passage through the ClpB pore.
Using high-resolution assessment, the researchers detected that step-sizes dictated the ClpB translocation modes, rather than step frequencies. They observed that slower runs established translocation steps of 14.6 amino acids, while faster ones established steps of 28 amino acids – an approximately two-fold difference.
Although the findings highlight how ClpB processes proteins and loop extrusion, the authors also suggest that the same type of regulated activity may extend to other polypeptide-processing enzymes.
For more information, you can read the full article entitled “Processive extrusion of polypeptide loops by a Hsp100 disaggregase” in the journal Nature.
Are you interested in using dynamic single-molecule tools like the C-Trap® for your research? Please feel free to contact us for more information, a demo, or a quote.