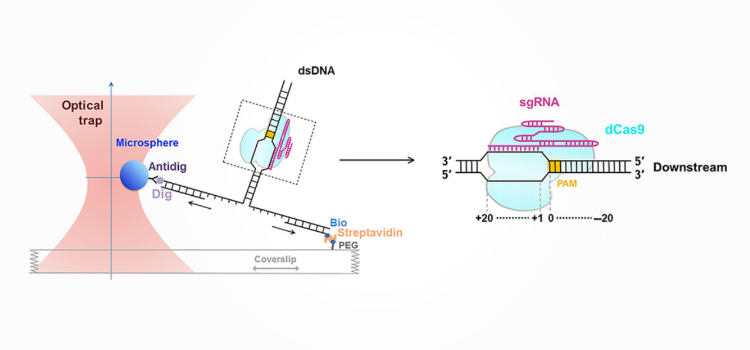
A new publication shows how the lab of Dr. Bo Sun used the m-Trap® to analyze the interactions between a Cas9–gRNA complex and DNA. Using a single-molecule approach to unzip the target double-stranded DNA, the team showed interaction points on the DNA that affected Cas9 binding and cleavage.
These results can help scientists find specific approaches to improve the gene-editing effectiveness of Cas9 by regulating the binding strength of the enzyme. Congratulations to all the authors!
Zhang et al. used high-resolution optical tweezers to unzip double-stranded DNA, a well-established technique that can measure the position and binding strength of proteins, such as Cas9, binding to a DNA molecule. The researchers found two interactions between the Cas9 complex and the DNA, flanking the protospacer adjacent motif (PAM).
One of these interactions localized fourteen base pairs downstream of the PAM and was weaker than the interactions measured in the gRNA-specific binding region. Interestingly, disruption of this so-called post-PAM interaction through unzipping collapsed the ternary formation between the Cas9-complex and the DNA.
Strong interactions and long dwell-times associated with Cas9–DNA complex formation could impede genome editing, for example by blocking DNA damage repair components. Hence the findings indicate that Cas9 activity may be optimized by regulating this critical interaction point downstream of the PAM.
For more information, you can read the full article entitled “The post-PAM interaction of RNA-guided spCas9 with DNA dictates its target binding and dissociation” in the journal Science Advances.
Related reading:
Are you interested in using dynamic single-molecule tools like the m-Trap® or C-Trap® for your research? Please feel free to contact us for more information, a demo, or a quote.