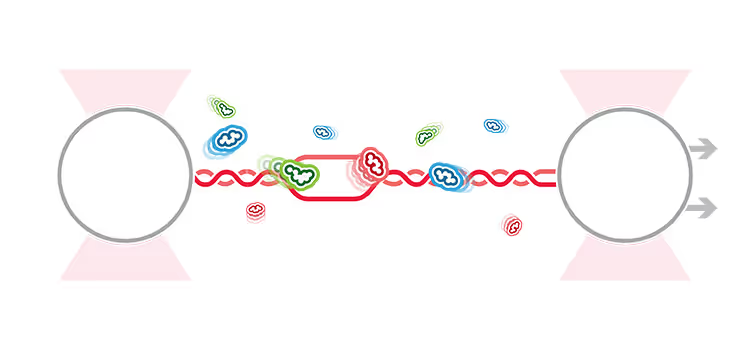
A recent study published in Molecular Cell used dynamic single-molecule analysis to evaluate the roles of specific proteins involved in the initiating steps of homologous recombination, including nucleation and growth of RAD51 filaments. Specifically, the researchers used the C-Trap® optical tweezers with correlated confocal microscopy to evaluate the roles of RAD51 paralogues and BRCA2 ortholog BRC-2 (mediator proteins) in stimulating RAD51-mediated processing of single-stranded DNA.
The study offers new insight into the initiation processes and regulation of homologous recombination. It focuses explicitly on how recombination mediator proteins orchestrate the displacement replication protein A (RPA) and stimulation of RAD51-filament growth.
To study these processes, Belan et al. first captured a double-stranded DNA between two optically trapped beads and produced single-stranded DNA (ssDNA) by stretching the substrate until the strands melted. The researchers followed and localized interacting proteins through the confocal microscope while moving the DNA substrate between protein channels of the C-Trap’s microfluidics system containing RAD51 or mediator proteins. With this setup, they could record how the relevant proteins localized and processed RPA-coated ssDNA in different conditions by fluorescently tagging specific proteins.
The team observed differential roles of the measured mediator proteins. For instance, they found that BRC-2 acts as a nucleation factor that displaces RPA and stimulates the assembly rate of RAD51 on the ssDNA. By contrast, they showed that Rad51 C. elegans paralog complex RFS-1/RIP-1 primarily stimulates RAD51-filament growth, specifically accumulating at the 5’-filament end and favoring asymmetric filament growth (3’ to 5’).
Interestingly, single-molecule analyses of mutant and wild-type forms of RFS-1/RIP-1 showed that these RAD51 paralogs dynamically associate with the formed filaments with high turnover. The authors suggest that this rapid turnover promotes filament growth in an ATPase-dependent manner and prevents disassembly.
The latter contrasts previous bulk electron microscopy (EM) data, suggesting that Rad51 paralogs stabilize Rad51 filaments and protects them from disruption through direct intercalation. The new insights emphasize the advantages of investigating dynamic and highly transient processes through single-molecule analysis and imaging in real time.
Read the full article titled “Single-molecule analysis reveals cooperative stimulation of Rad51 filament nucleation and growth by mediator proteins” on Molecular Cell to find out more about these findings.