Chromatin structure and remodeling is a fundamental process that ensures the dynamic regulation of DNA accessibility in our cells. This process is crucial for essential functions such as gene expression, DNA replication and repair. Chromatin remodelers, the protein complexes responsible for these adjustments, play a pivotal role in regulating gene accessibility. Misregulation of these processes can lead to a variety of diseases, including cancer.
Chromatin remodelers are known to move along DNA, reposition nucleosomes, and alter histone modifications, which affects chromatin compaction and accessibility. These remodelers can also exchange histone variants and interact with other chromatin-associated proteins to modulate gene expression. However, the precise mechanism by which remodelers search, engage and direct nucleosome translocation, has remained largely unknown.
Recently, Prof. Taekjip Ha and Prof. Carl Wu from Johns Hopkins University and Howard Hughes Medical Institute have made a groundbreaking discovery, revealing that chromatin remodelers can employ various mechanisms when interacting with DNA and nucleosomes. Utilizing cutting-edge dynamic single-molecule analysis, these researchers provided the first real-time, detailed observation of these dynamic processes at the single-molecule level. The results of this study were published in the journal eLife.
Chromatin remodeler protein complexes
SWI/SNF (Switch/Sucrose Non-Fermentable): This family of remodelers is known for its role in enhancing gene expression by making DNA more accessible. They can slide nucleosomes along the DNA or eject them entirely, thus opening regions of the DNA for transcription. Mutations in these proteins are often associated with cancer.
ISWI (Imitation Switch): ISWI remodelers typically slide nucleosomes along DNA without ejecting them, maintaining a more orderly chromatin structure. These proteins are crucial for processes like DNA replication and repair, and gene expression regulation.
CHD (Chromodomain Helicase DNA-binding): Members of the CHD family are characterized by the presence of chromodomains that can bind to methylated DNA or histones, influencing their activity. These remodelers are involved in both gene activation and repression, depending on the context and specific CHD protein involved.
INO80 (INO80 Complex): The INO80 family can both slide nucleosomes and facilitate histone exchange. These remodelers are particularly important in DNA repair and may play a role in replication.
Dynamic Mechanisms of Chromatin Remodelers: RSC and ISW2
Chromatin remodeler RSC and ISW2 are pivotal in regulating gene transcription and chromatin structure through their distinct mechanism of action. RSC (from the SWI/SNF family) promotes chromatin opening, increasing DNA accessibility for transcription, while ISW2 (from the ISWI family) compacts chromatin, thereby reducing accessibility. This contrasting regulation mechanism of chromatin accessibility is crucial for controlling gene transcription.
Despite extensive research using live-cell fluorescent single-particle tracking, the exact mode of dynamic translocation of RSC and ISW2 remained unknown due to the crowded environment of the nucleus and the inability to observe individual remodelers in action clearly. Key questions in the field include whether there are differences in their opposing directional movement and what happens during remodeler-remodeler or remodeler-nucleosome collisions.
Using LUMICKS’ C-Trap, an instrument that combines single-molecule manipulation with fluorescence microscopy, the authors overcame these challenges. Chromatin remodelers RSC and ISW2 were visualized in real-time and with single-molecule resolution as they translocated, repositioned nucleosomes, moved along DNA, and formed transient complexes on DNA.
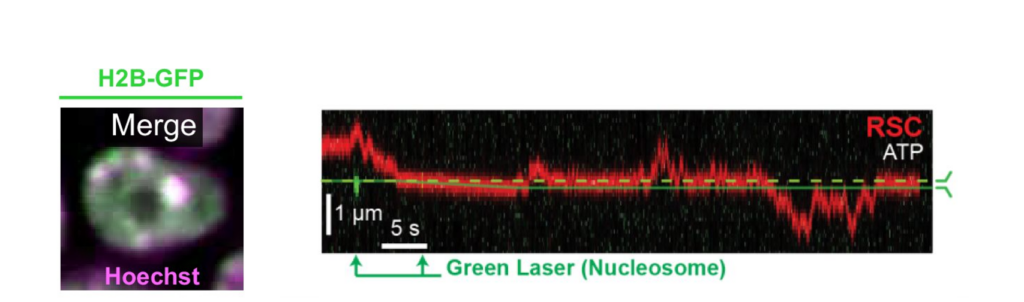
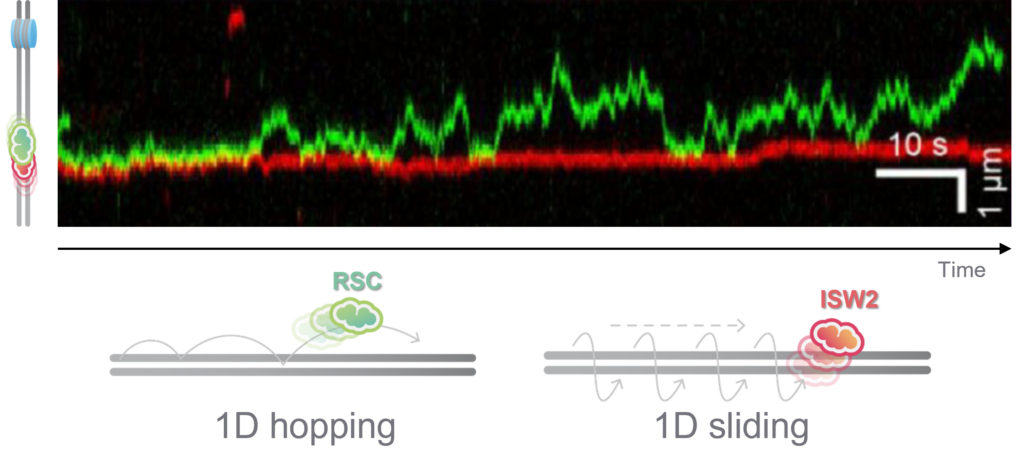
Interestingly, RSC although a much larger protein complex, displayed a higher mobility along DNA compared to ISW2, suggesting different DNA binding mechanisms for translocation along DNA. Based on their observation, the scientists could confirm that RSC utilizes a ‘hopping’ mechanism for translocation, which is more dynamic under varying ionic conditions, while ISW2 employs a ‘sliding’ mechanism, maintaining consistent contact on DNA regardless of ion concentration (Figure 1). These distinct translocation strategies reflect different functional roles in chromatin remodeling.
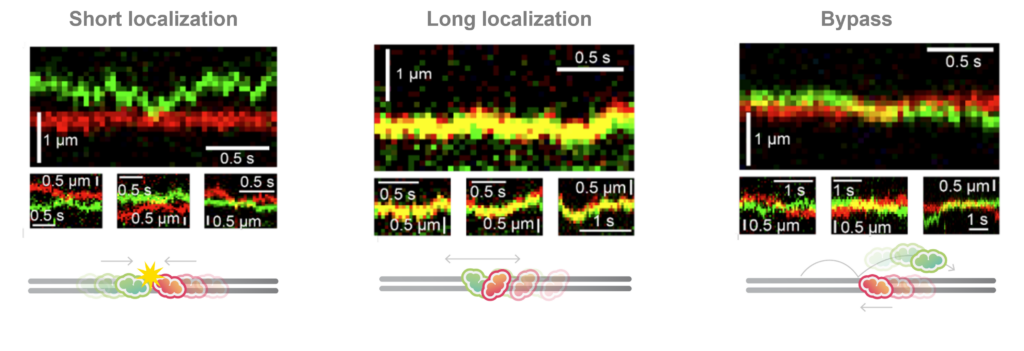
What happens when the chromatin remodelers meet each other? With multi-color fluorescence imaging, the authors discovered three different types of interactions: short-lived colocalization, long-lived colocalization and bypassing (Figure 2). These behaviors suggest the chromatin remodelers function as roadblocks for each other and have mutual affinity while bound on the DNA at the same time.
Pushing and pulling nucleosomes
As remodelers usually function by repositioning nucleosomes on the DNA, it’s of great importance to directly visualize the dynamics of chromatin remodelers encountering nucleosomes at the single-molecule level. Researchers have found that in the absence of ATP, the energy molecule that powers chromatin remodeling, remodelers tend to colocalize with and bypass nucleosomes. However, with ATP present, remodelers are capable of actively translocating nucleosomes for up to 4000 bp.
Single-molecule observations also showed distinct behaviors between different remodelers: RSC tends to move nucleosomes in a ‘pushing-like’ fashion, while ISW2 moves them in the opposite direction, thereby ‘pulling’ them. These findings, illustrated in Figure 3, provide the first direct visual evidence validating the push-pull model, which describes how remodelers can expand or contract nucleosome-free areas. This insight into chromatin dynamics is pivotal for understanding the regulation of gene access and expression.

Significance and implications
Chromatin remodelers are essential as they play a central role in regulating gene expression and maintaining genomic integrity, both of which are crucial for proper cellular function and the overall health of organisms.
Using the C-Trap, researchers have for the first time directly observed two distinct behaviors of chromatin remodelers: ‘hopping’ and ‘sliding’ along DNA. Besides, this publication provided invaluable insights into the remodeler’s dynamic actions, including a ‘pushing and pulling’ model of chromatin remodeling.
These findings, made possible exclusively through the use of the C-Trap, not only deepen our understanding of chromatin dynamics but also lay the groundwork for future scientific breakthroughs in Chromatin dynamics research.
For further information, read the full article “Dynamic 1D Search and Processive Nucleosome Translocations by RSC and ISW2 Chromatin Remodelers”, published in Life.
Are you interested in using dynamic single-molecule tools like the C-Trap for your research?
For more information, a demo, or a quote contact us!
[1] May D, Yun S, Gonzalez DG, Park S, Chen Y, Lathrop E, Cai B, Xin T, Zhao H, Wang S, Gonzalez LE. Live imaging reveals chromatin compaction transitions and dynamic transcriptional bursting during stem cell differentiation in vivo. Elife. 2023 Mar 7;12:e83444.
[2] Kim JM, Carcamo CC, Jazani S, Xie Z, Feng XA, Yamadi M, Poyton M, Holland KL, Grimm JB, Lavis LD, Ha T. Dynamic 1D search and processive nucleosome translocations by RSC and ISW2 chromatin remodelers. Elife. 2024 Mar 18;12:RP91433.



