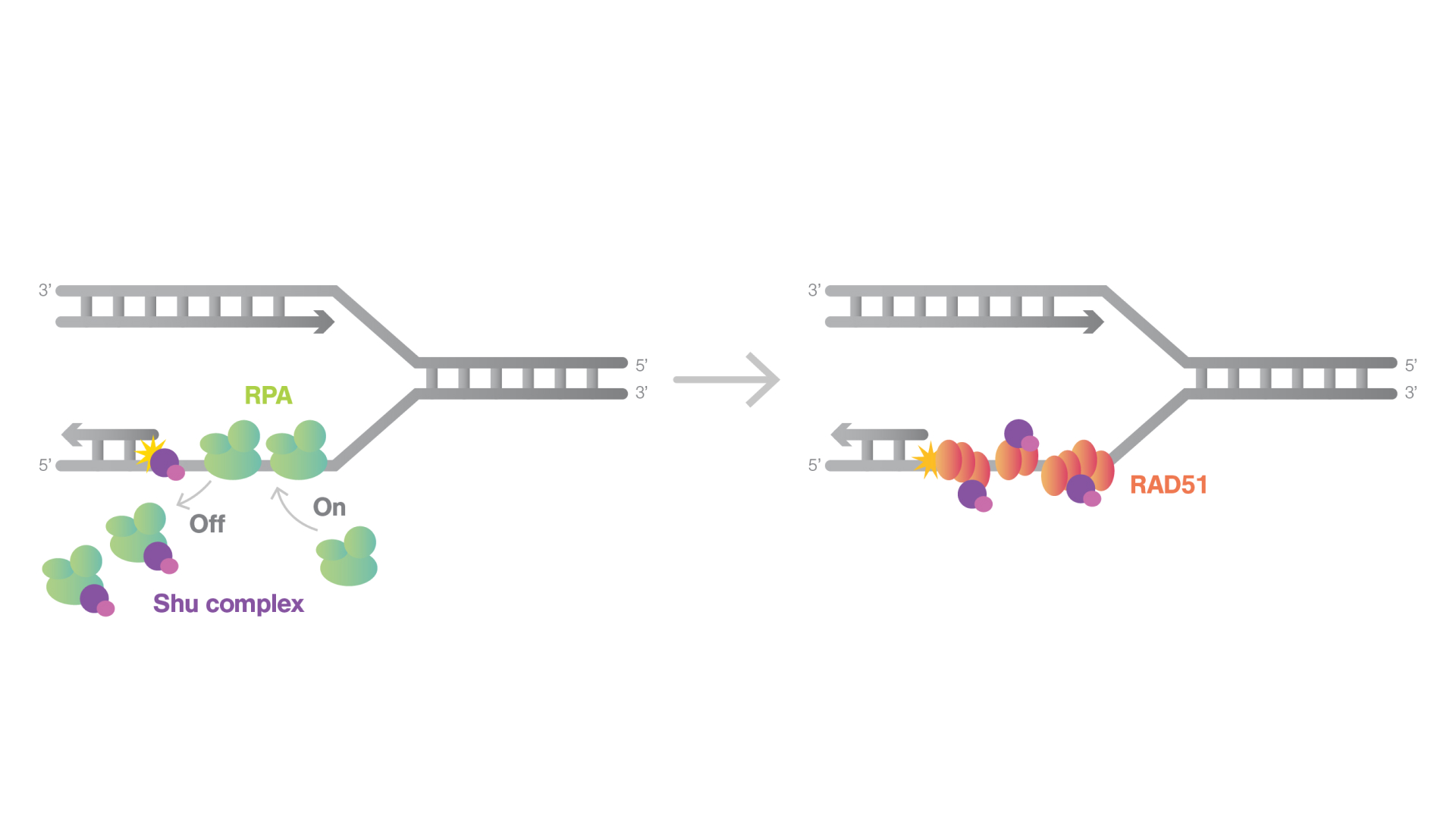
Homologous recombination is a vital cellular process for repairing DNA double-strand breaks, ensuring both the repair and proper segregation of chromosomes during cell division. Central to this mechanism is the activity of RAD51 filaments, which is critically supported by the Shu complex. Shu complex is pivotal in DNA damage repair and consists of RAD51 paralogs. Mutations in these paralogs are linked to cancer predisposition and Fanconi anemia. Like most RAD51 paralogs, the Shu complex interacts with Rad51 to influence Rad51 filament assembly, crucial for the repair process.
Despite over 15 years of research, the precise mechanistic function of the Shu complex in RAD51-dependent repair remained obscure. The role of the human Shu complex in DNA repair had been primarily inferred from yeast studies, with limited direct evidence of its function in human cells, particularly regarding its interactions with RAD51 and RPA on single-stranded DNA (ssDNA).
Important proteins in homologous recombination
RAD51 – Central to homologous recombination, RAD51 forms nucleoprotein filaments necessary for homology search and DNA strand invasion, directly facilitating the repair of double-strand breaks by promoting strand exchange between homologous DNA strands.
RAD52 – Assists in the loading of RAD51 onto single-stranded DNA and also plays a role in the annealing of complementary DNA strands, serving as a mediator in the recombination process.
RAD54 – Works synergistically with RAD51, promoting DNA strand exchange by remodeling DNA and chromatin structures around the site of damage, enhancing the accessibility of the DNA to repair proteins.
RPA (Replication Protein A) – Binds to exposed single-stranded DNA at resection sites, protecting it from degradation and helping to properly align the DNA for the recruitment of RAD51 and other recombination proteins.
BRCA1 and BRCA2 – These tumor suppressor proteins are crucial for the regulation of RAD51, ensuring its proper function and localization during DNA repair. BRCA1 also interacts with several other repair proteins to facilitate efficient repair, while BRCA2 directly binds to RAD51, modulating its filament formation.
DMC1 – This meiosis-specific recombinase facilitates homologous chromosome pairing and strand exchange during meiosis, functioning similarly to RAD51 but with a specific role in germ cells.
MSH2 and MSH6 – These are components of the mismatch repair system but are also involved in signaling and processing DNA breaks for homologous recombination, particularly in response to replication stress.
XRCC3 – A RAD51 paralog that plays a role in stabilizing RAD51 complexes and helps maintain chromosome integrity during cell division, particularly important for the repair of cross-links and double-strand breaks.
PALB2 – Functions as a partner and localizer of BRCA2 in DNA repair; it is essential for recruiting BRCA2 and RAD51 to sites of DNA damage, thereby playing a key role in the repair mechanism.
CTIP (also known as RBBP8) – Critical for the initial step of resecting DNA double-strand breaks to create single-stranded DNA tails. CTIP facilitates the recruitment and loading of the recombination machinery by modulating DNA end resection.
Recent breakthroughs have shed light on these mechanisms. Professors Sarah Hengel from Tufts University and Prof. Kara Bernstein from University of Pennsylvania School of Medicine published their findings in Nature Communications, revealing new insights into the Shu complex’s function with RAD51 filaments using C-Trap Dynamic single-molecule microscope. C-Trap is the only technology that allows large RAD51 filaments to be visualized in a controlled environment, providing a unique view into the dynamics of DNA repair.
In this innovative study, the researchers utilized biotinylated ssDNA(20,452nt), specifically engineered for use with the C-Trap system. This kit enabled protein binding experiments on ssDNA. The researchers could quickly move the ssDNA across different microfluidic channels containing fluorescently labeled RAD51, Shu complex, RPA, or various combinations of these proteins. The rapid transition between channels allowed for quick alterations in the buffer conditions, facilitating dynamic experiments.
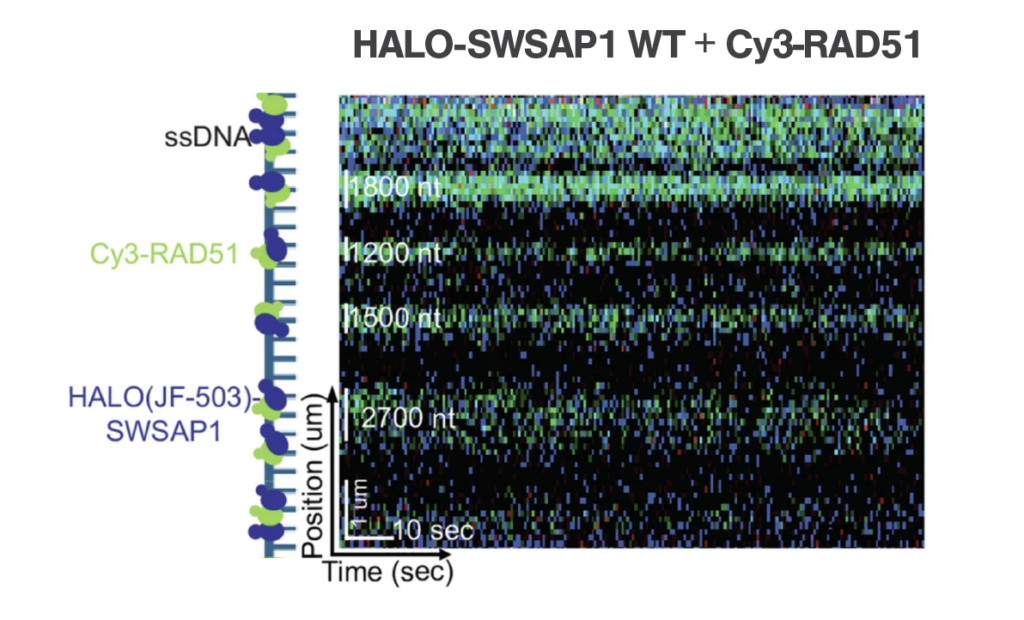
Unlike other RAD51 paralogs that typically cap the filament ends, the authors found that Shu complex was predominantly colocalized along the RAD51 filaments on ssDNA. This indicates that the Shu complex may enhance the stability of RAD51 filaments, potentially by increasing nucleation sites or altering the conformation of RAD51 on the ssDNA.
For the first time, the authors discovered that the Shu complex significantly improves the diffusion of RPA on ssDNA. During homologous recombination, RAD51 filaments displace RPA from ssDNA to initiate repair. The findings suggest that the Shu complex increases the mobility of RPA on ssDNA, thereby accelerating its dissociation. This action promotes the formation of RAD51 filaments, thus enhancing the efficiency of DNA repair.
Building on their findings from experiments with knockout and cancer cell models, the authors concluded that the Shu complex plays a crucial role in stimulating high-fidelity RAD51-dependent DNA repair. This significant discovery positions the Shu complex as a potentially vital target for cancer therapy.
The impact of this research extends beyond basic biological understanding, suggesting promising new avenues for the development of targeted cancer treatments that could enhance the precision of DNA repair mechanisms, crucial for cancer prevention and therapy.
Furthermore, the methods employed, particularly using the C-Trap system to visualize protein interactions on ssDNA, demonstrate a versatile approach that can be adopted by further research in DNA repair studies.
Images are adapted from the original publication: Hengel, Sarah R., et al. “The human Shu complex promotes RAD51 activity by modulating RPA dynamics on ssDNA.” Nature communications 15.1 (2024): 7197.