A new approach that enables the full understanding of molecular mechanisms
Biological processes occur at the molecular level, where diseases originate and the majority of drugs act. Thus, understanding the details of molecular mechanisms like DNA damage response, is crucial.
Existing life science tools (such as structural biology, bulk functional assays, cell imaging, and localization assays) offer detailed structural or dynamic functional data, but never both.
Dynamic single-molecule microscopy fills this gap by revealing intricate mechanistic details of protein-DNA interactions in real-time
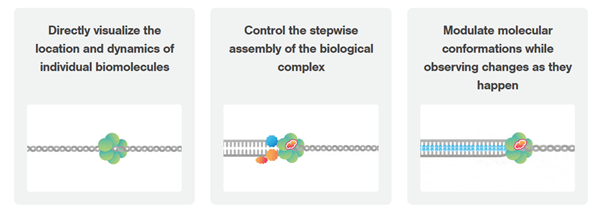
The exact order and timing of molecular events is inaccessible with established biochemistry assays?
Characterize the (dis-)assembly kinetics of repair complexes based on single-molecule real-time data
Understanding the sequence of events in assembling multi-protein complexes is crucial for comprehending DNA repair processes and identifying potential drug targets.
LUMICKS’ dynamic single-molecule approach provides such insights, even within the physiological environment of cellular extracts.
Professor Ben Van Houten‘s team at the University of Pittsburgh successfully used cellular extracts without complex protein purification to
- track multi-step binding processes
- quantify binding kinetics
- reveal molecular mechanism heterogeneity in UV-DDB-related DNA repair processes
Use the force: Furthermore, using the C-Trap’s ability to apply precise mechanical stress on individual molecules, they discovered a correlation between PARP1 binding kinetics to nicks, and the mechanical state of the DNA substrate.
- Jang et al. Nucleic Acids Research (2022)
- Wan et al. PNAS (2023)
- Schaich et al. Nucleic Acids Research (2023)
- Schaich et al. DNA Repair (2024)
Watch our Nature webinar!
Single-molecule analysis of cancer DNA-protein interactions from nuclear extracts
Dr. Bennet Van Houten and his team developed a new technique performing dynamic single-molecule analysis directly on nuclear extracts, without the need for purification of recombinant protein. This allows rapid mechanistic analysis of mutant proteins from cancer cells, providing previously unseen insights into their mechanisms of action.
ADD TESTIMONIAL

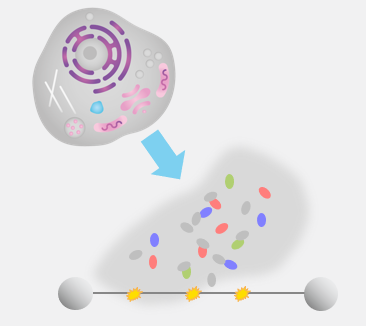
Step 1: Proteins are expressed in cells and nuclear extracts are loaded into the C-Trap where they interact with well-defined DNA substrates.
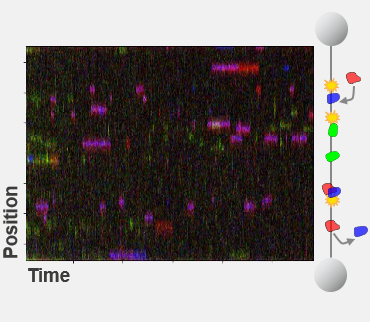
Step 2: Single-molecule fluorescence imaging allows the identification and real-time tracking of multiple types of proteins on the DNA substrate.
Step 3: Easy-to-use data analysis provides insights into the steps, kinetics, and potential variations of the molecular process.
Lacking the full picture of regulative elements in complex molecular mechanisms?
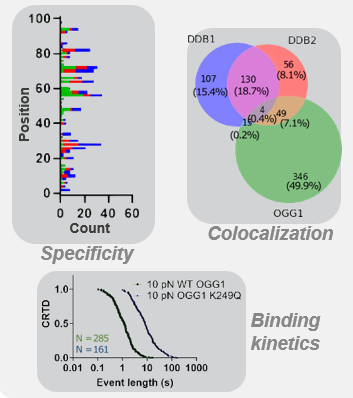
Quantify the activity of selected components in DNA repair processes
Homologous recombination (HR) is crucial for repairing double-strand breaks (DSBs) in DNA, yet some molecular mechanisms and protein roles remain unclear.
Utilizing the C-Trap, studies led by Steve West, Simon Boulton (both at The Francis Crick Institute, London), and David Rueda (Imperial College, London) uncovered new sub-steps and regulatory functions in RAD51 filament formation, a key HR process:
- Real-time observations revealed BRCA2’s role in nucleating and stabilizing RAD51 on RPA-coated ssDNA
- A diffusion-assisted mechanism involving dsDNA binding and sliding was identified
- Additionally, RFS-1/RIP-1 was found to act as a dynamic ‘chaperone’ to promote filament growth
- BCDX2 in turn was found to stimulate RAD51 filament nucleation and extension, dependent on RAD51B and RAD51C ATPase activities.
These findings elucidate two distinct BRCA2-dependent RAD51 loading mechanisms onto ssDNA and illuminate the roles of recombination mediators during filament growth.
Understanding these processes at the single-molecule level clarifies the function of BCDX2 and other factors in RAD51 assembly on ssDNA, which is crucial for replication fork protection and DSB repair, and essential for tumor avoidance.
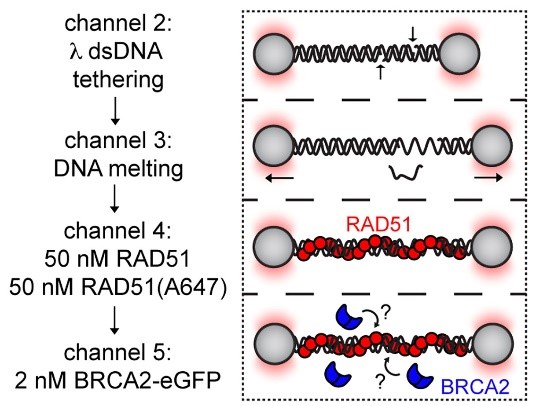
Schematic of RAD51 filament binding experiment, in which λ DNA was pre-incubated with a 1:1 mixture of labeled and unlabeled RAD51 and then moved to a channel containing BRCA2-eGFP to monitor BRCA2 binding.
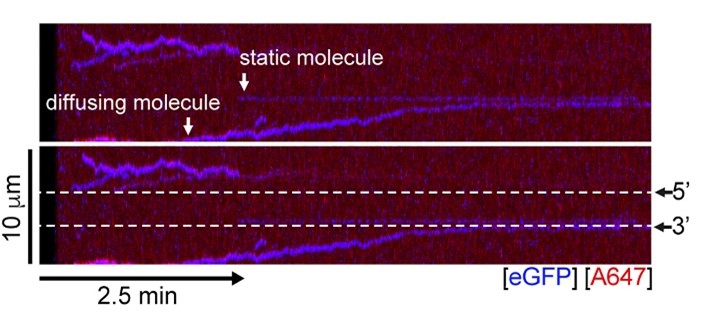
A representative kymograph showing diffusion-driven delivery of BRCA2-RAD51 complexes to ssDNA in the vicinity of the ds-ssDNA junction. Static BRCA2-eGFP molecules bound directly to the ssDNA gap. Mobile BRCA2-eGFP molecules diffuse along dsDNA. 25 nM RAD51-A466 (red) was incubated with DNA in the presence of 5 nM BRCA2-eGFP (blue) and 1.25 nM RPA in the presence of 5 nM SYTOX Orange. Position of the ssDNA gap is indicated by dashed lines.
Missing unambiguous evidence of your new hypothesis?
Directly observing molecular search and repair mechanisms delivers unexpected insights
Static structural data often limits hypotheses and models explaining DNA-protein binding mechanisms, failing to capture complex dynamics.
Ingrid Tessmer (Rudolf Virchow Centre Würzburg) and her team employed the C-Trap to directly visualize DNA translocation and lesion recognition by O6-alkylguanine DNA alkyl-transferase (AGT).
This approach unveiled details of AGT’s capabilities to
- dynamically bind DNA
- form clusters
- search lesions
Real-time observation with the C-Trap provided quantitative insights, challenging established models of AGT’s unidirectional movement, previously believed to be accelerated by cluster formation.
Not fully confident about the mode of action of potential anti-cancer drugs?
Understand small molecule inhibitors by watching them in action
Double-stranded breaks (DSBs) in DNA pose significant threats, contributing to cell death and cancer. POLQ, an enzyme implicated in DSB repair, is often overexpressed in cancers, bolstering cancer cell survival.
Researchers at The Francis Crick Institute, led by Simon Boulton, utilized the single-molecule imaging capacity of the C-Trap to
- measure DNA polymerase theta (POLQ) efficiency
- observe its role in sealing post-replicative single-stranded DNA (ssDNA) gaps
- directly visualize and quantify the inhibiting effects of POLQ targeting drugs
This discovery aligns with previous findings showing a synergistic effect between POLQ and PARP inhibitors, a crucial therapy for BRCA-deficient cancers.
Simon Boulton’s team suggests that POLQ inhibitors could serve as potent second-generation drugs for treating PARP-inhibitor-resistant BRCA-deficient cancers, shedding light on potential novel treatment avenues.
Learn more and watch a 60s summary of the key findings!
Wondering how molecular interactions physically impact DNA dynamics?
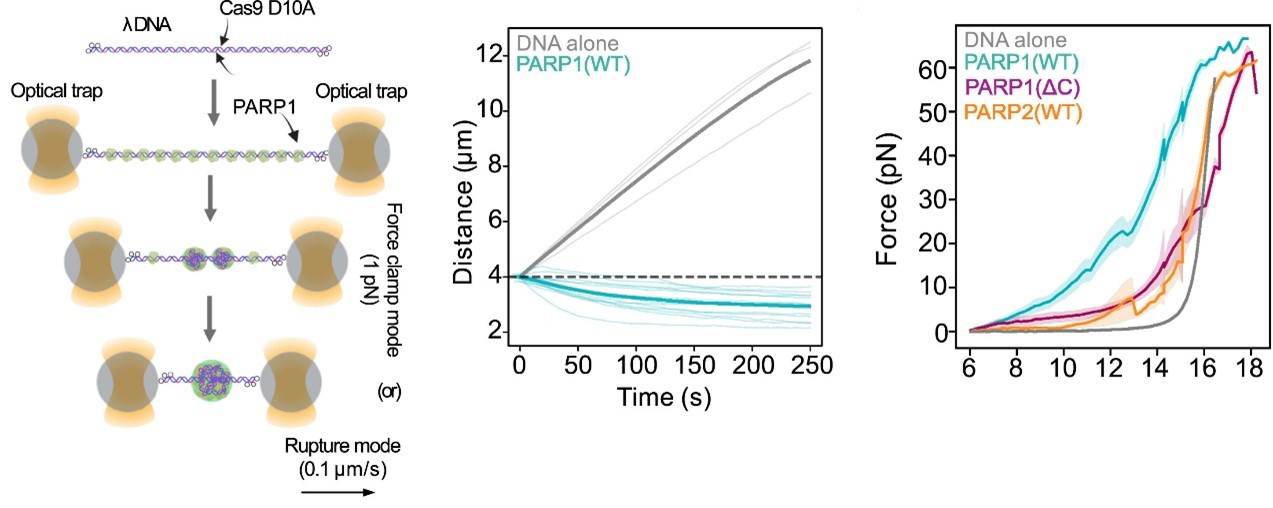
Left: Workflow of the optical tweezer force clamp and force-extension experiments.
Center: Distance measurement as a function of time from force clamped λ DNA or λ DNA harboring a PARP1 condensate. Thick line represents mean value.
Right: Force measurement as a function of distance from tethered λ DNA extended in the presence of mEGFP-tagged PARP1(WT) or PARP1(ΔC) or PARP2(WT) (1 μM). Thick line represents the mean value.
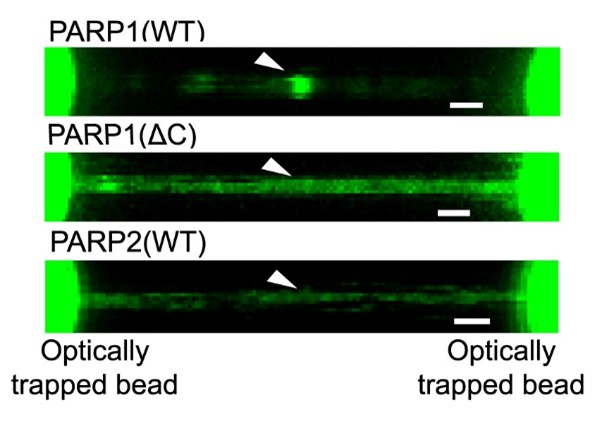
Representative florescence images of mEGFP-tagged PARP1(WT), PARP1(ΔC), and PARP2(WT) localization on tethered λ DNA, which was nicked on both strands by CAS9(D10A) with a 60 bp gap between the nicks. Only PARP1(WT) shows condensation at the damage site. Scale bar, 2 μm.
Images from: Chappidi et al. Cell 2024
Use the force: Pulling on individual molecules reveals how biomolecular condensation physically prevents DNA end disjunction
Preventing DNA end disjunction is vital for DNA double-strand break (DSB) repair, yet the role of PARP1 in this process remains unclear.
Simon Alberti (MPI-CBG Dresden) and collaborators used LUMICKS’ dynamic single-molecule approach with the C-Trap to investigate PARP1 recruitment to DSBs.
Single-molecule imaging, combined with measuring mechanical forces, revealed PARP1’s formation of condensates around damage sites.
These biomolecular condensates physically connect DNA ends, even under tension, facilitating the recruitment of additional repair and regulatory factors.
Visit our blog post to learn more about the study and the methods used!
Our solutions
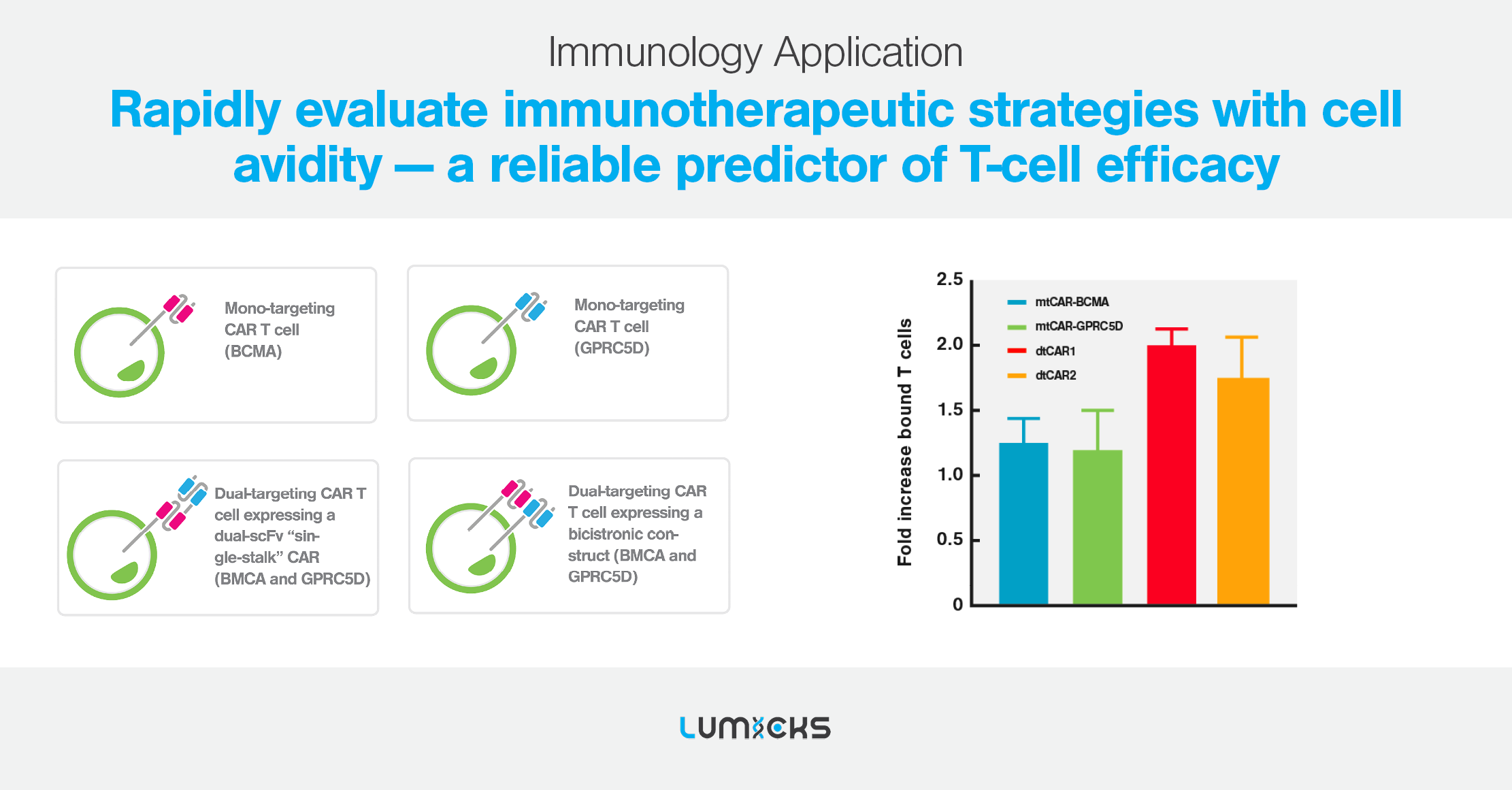
Cell avidity as a service
Cell Avidity analysis can be performed at high throughput using our services platform, enabling the screening of hundreds of CAR-T cells to rapidly find the most potent CAR-T for in vivo testing.
Dr. Rogier Reijmers
Principal Scientist at LUMICKS