MeCP2, a crucial epigenetic regulator involved in the neurodevelopmental disorder Rett Syndrome
Methyl-CpG-binding protein 2 (MeCP2) is a key regulator of gene expression, primarily known for its role in binding to methylated DNA at CpG sites to suppress gene activity. Mutations in MeCP2 lead to Rett syndrome, a neurodevelopmental disorder affecting mainly young females. Since its discovery in 1992, significant advancements have been made in understanding MeCP2’s function. The protein is known to interact with various regulatory proteins in different contexts, influencing transcription. However, the exact mechanisms allowing MeCP2 to fulfill its various roles remain elusive.
MeCP2’s highly disordered structure has posed challenges to traditional structural studies. Techniques like proteomics and biochemical assays only provide indirect insights into MeCP2’s activity. To overcome these limitations, Dr. Shixin Liu’s team at Rockefeller University utilized the C-Trap, a dynamic single-molecule microscope. This approach enabled them to explore the dynamics and kinetics of MeCP2, including its interactions with chromatin and the effects of disease-related mutations.
Key Concepts and Proteins in Epigenetic Regulation:
Epigenetic regulation involves several mechanisms that alter gene expression without changing the underlying DNA sequence. These modifications play critical roles in cellular differentiation, development, and the response to environmental factors. Below are key concepts and proteins central to epigenetic regulation:
DNA Methylation: Addition of methyl groups to DNA, typically at CpG sites, leading to gene suppression. Proteins like DNMT1 (DNA methyltransferase 1) are responsible for maintaining DNA methylation patterns during cell division, crucial for proper gene repression.
RNA Methylation: This involves the methylation of RNA molecules, especially at the N6-methyladenosine (m6A) site, impacting RNA stability and translation efficiency. Proteins like METTL3 are critical m6A writers, while FTO functions as a demethylase, regulating RNA methylation dynamics.
Histone Modification: Histones can be chemically modified by methylation, acetylation, phosphorylation, and ubiquitination, influencing chromatin structure and gene expression. Proteins like histone deacetylases (HDACs) remove acetyl groups from histones, leading to tighter chromatin packing and gene repression. Conversely, histone acetyltransferases (HATs) promote transcriptional activation by adding acetyl groups.
Chromatin Remodeling: This involves altering the structure of chromatin to make DNA more or less accessible to transcription factors. Complexes like SWI/SNF remodel chromatin by sliding nucleosomes along DNA, facilitating transcriptional activation or repression depending on context.
Non-Coding RNA (ncRNA) Interaction: nteraction: ncRNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), modulate gene expression post-transcriptionally or by altering chromatin states. For example, Xist, a lncRNA, plays a critical role in X-chromosome inactivation by recruiting chromatin modifiers to silence one X chromosome in females.
Visualization of MeCP2 Dynamics Using LUMICKS Single-Molecule Assay
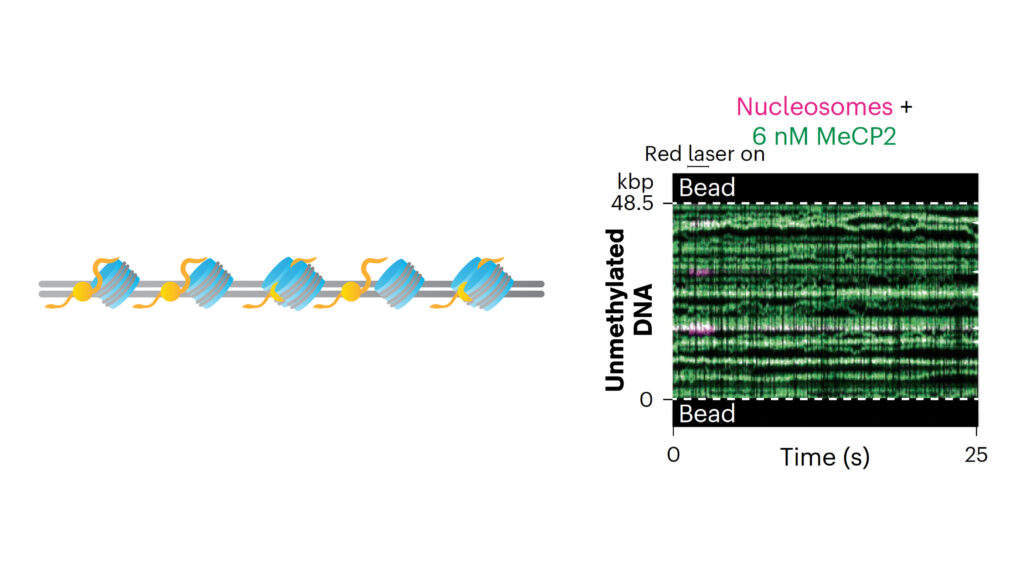
The Rockefeller University’s researchers utilized the dual optical traps of the C-Trap to suspend a single DNA molecule in solution and introduced purified human MeCP2. This setup enabled the real-time observation of MeCP2’s movement along DNA at the single-molecule level. Additionally, they utilized the advanced microfluidics system of the C-Trap to add multiple proteins such as histone octamers and chromatin regulators, enabling the observation of MeCP2’s interactions with chromatinized DNA and its coordination with other transcriptional regulators.
Discovery and Implications of the Research
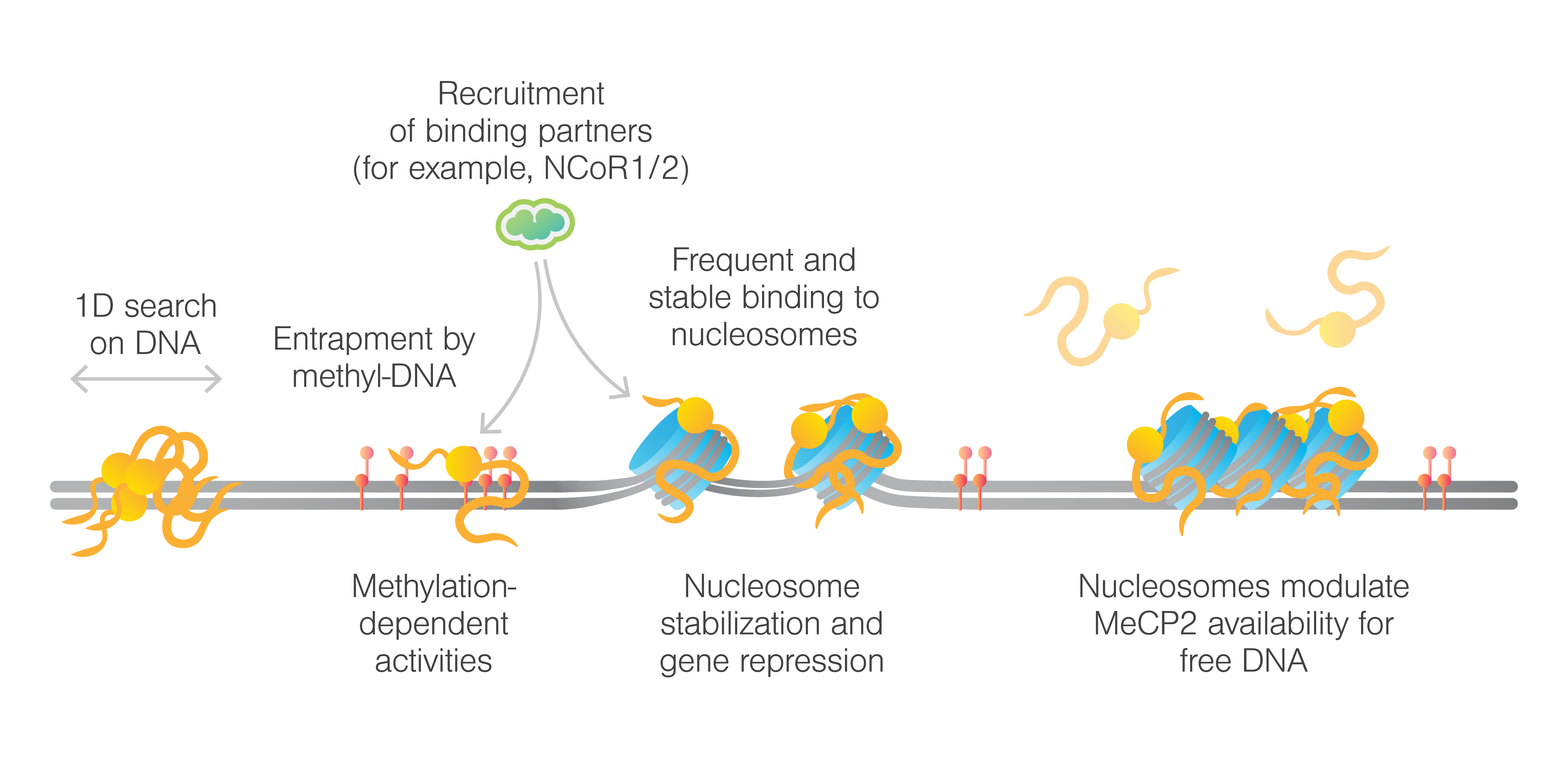
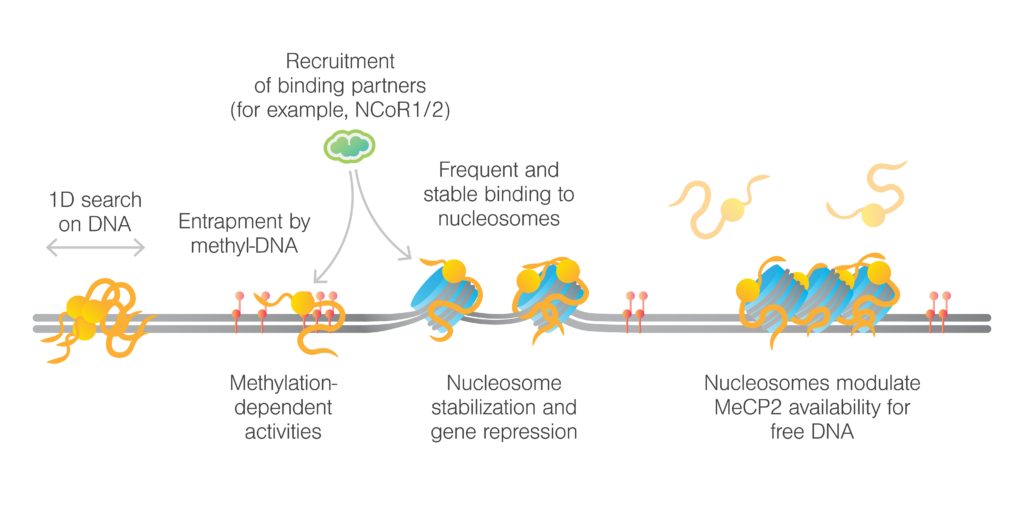
The study demonstrated, for the first time, that MeCP2 can slide on unmethylated λ DNA, whereas its movement is significantly slower on methylated DNA. The researchers also examined various RTT mutations in MeCP2, discovering that these mutations distinctly affect both the protein’s mobility on DNA as well as its binding ability. Furthermore, measurements with chromatinized DNA showed that MeCP2 remains stationary at nucleosome sites on both methylated and unmethylated DNA. Truncation experiments highlighted the essential role of the TRD (transcriptional repression domain) in targeting nucleosomes. Furthermore, mechanical pulling on chromatinized DNA with bound MeCP2 demonstrated that a higher force was required to disrupt nucleosome structures, indicative of a stabilizing, effect reducing chromatin accessibility.
Additionally, the study explored MeCP2’s interactions with chromatin effector proteins, such as the NCoR1/2 co-repressor complex, which is known to interact with MeCP2 via the transducin β-like protein 1-related (TBLR1) component. The team also discovered that the R306C mutation, common in Rett syndrome, severely impairs MeCP2’s ability to recruit TBLR1, a co-repressor complex crucial for regulating gene expression. This finding is particularly significant as it highlights a fundamental disruption in the molecular machinery that differentiates healthy cells from diseased ones.
Conclusion
This study establishes a robust framework for analyzing the unusual behaviors of MeCP2 in healthy and disease mutants in individuals with Rett syndrome (RTT). Given the challenge of limited structural data, examining MeCP2’s biophysical interactions with DNA and other regulatory complexes at the single-molecule level is crucial for understanding its mechanisms. This research delivers valuable molecular insights into MeCP2 and RTT, emphasizing the crucial role of nucleosomes in shaping the chromatin. These findings hold promise for developing targeted therapeutics aimed at correcting the molecular disruptions caused by MeCP2 mutations.
Images are adapted from the original publication: Chua, G.N.L., Watters, J.W., Olinares, P.D.B. et al. Differential dynamics specify MeCP2 function at nucleosomes and methylated DNA. Nat Struct Mol Biol (2024). https://doi.org/10.1038/s41594-024-01373-9
Prof. Shixin Liu, the the corresponding author of the publication, has published 13 publications with an average impact factor of 14.7 in the past 5 years with the C-Trap.
We are excited to share that he is giving a live webinar with us in December. It’s a great chance to learn about this publication and his research on DNA and chromatin. Register now to secure your spot!