The maintenance of genomic stability is crucial for the prevention of cancer and other diseases. One of the key players in this process is the Fanconi anemia (FA) pathway, which is responsible for repairing DNA interstrand crosslinks (ICLs). Central to this pathway is the FANCD2-I (D2-I) complex, whose new mechanism has recently been elucidated, shedding light on its critical role in DNA repair.
Understanding the Fanconi Anemia Pathway
Fanconi anemia (FA) is a complex genetic disorder characterized by an inability to repair DNA interstrand cross-links (ICLs). The FA pathway functions within a critical tumor-suppressor network to preserve genomic integrity by stabilizing replication forks, mitigating replication stress, and regulating cytokinesis. Researchers have identified over 20 genes that regulate the FA DNA repair pathway. This pathway involves a series of intricate interactions among several proteins, with the D2-I complex playing a central role in orchestrating the repair process.
The Role of D2-I Complex
The D2-I complex consists of FANCD2 and FANCI proteins, which form a heterodimer to recognize and repair DNA damage. The D2-I complex binds to double-stranded (ds) DNA and is essential for recruiting downstream proteins. It also plays a significant role in maintaining DNA integrity during replication stress.
A key event in the FA DNA repair pathway is the monoubiquitination of the FANCI and FANCD2 proteins within the D2-I complex. The monoubiquitinated D2-I complex encircles DNA and stably localizes to lesions, facilitating the recruitment of other repair proteins to the site of damage, thereby initiating the repair process.
Numerous studies, including structural research, have revealed different functions of the D2-I complex, including detailed conformational changes upon ubiquitination. However, the molecular mechanism by which the FA pathway recognizes DNA crosslink sites and how the D2-I complex functions in replication fork protection remains unknown.
New Discoveries of D2-I Mechanism
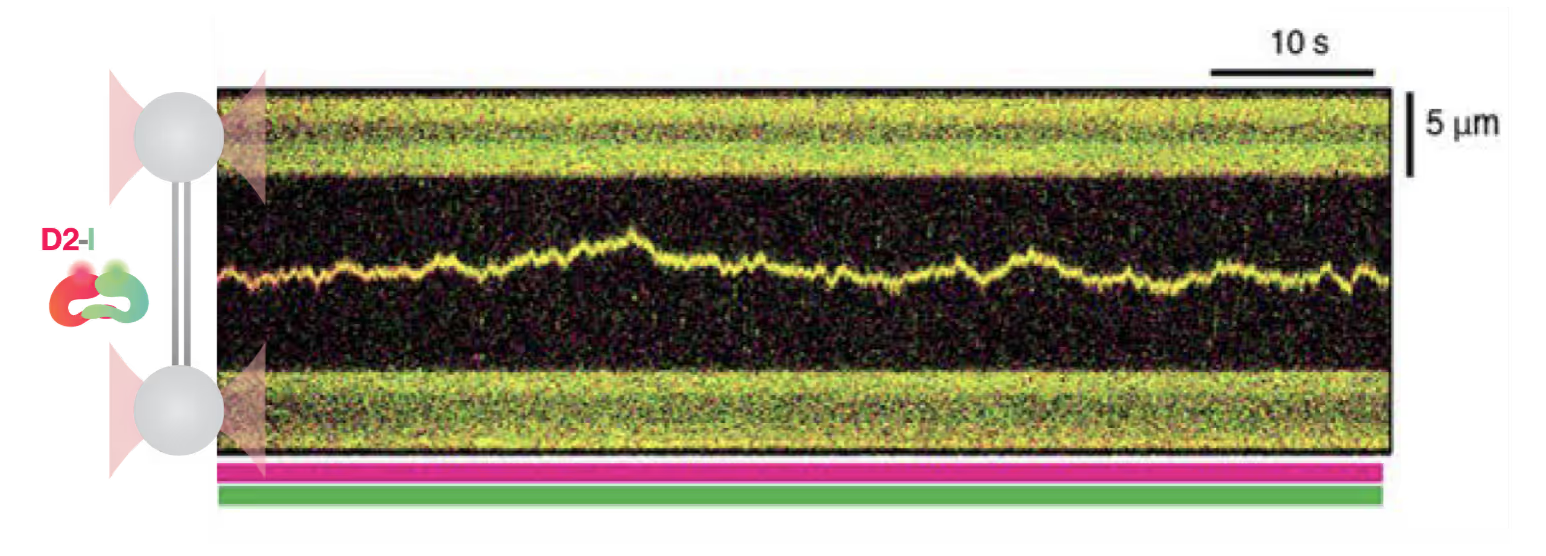
A recent Nature paper by the group of Prof. David Rueda (Imperial College London) and Prof. Lori Passmore (MRC Laboratory of Molecular Biology) has uncovered significant insights into the recognition of DNA crosslinks and the functional dynamics of the D2-I complex in replication fork protection. Using a dynamic single-molecule microscope, C-Trap, the researchers observed that the D2-I complex binds to and slides along double-stranded (ds) DNA but stalls at junctions between double-stranded and single-stranded (ds-ss) DNA. This finding elucidates the role of D2-I in recognizing and protecting stalled replication forks, offering new perspectives on DNA repair mechanisms and potential cancer therapies.

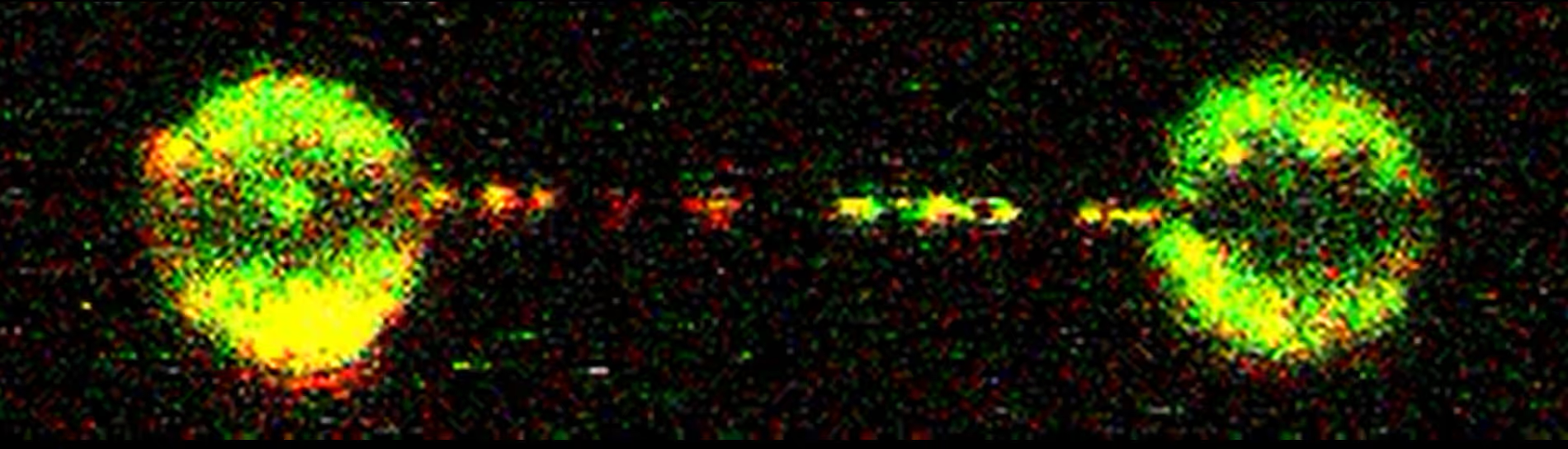
The study demonstrates that the D2-I heterodimer forms a clamp around the DNA, allowing it to slide. With the C-Trap, the authors achieved single-molecule resolution in real-time, tracking the movement of the D2-I complex along the DNA. The recorded kymographs show that the D2-I complex moves at a diffusion speed of 0.81 µm/s, and a long residence time on DNA of 119 seconds, indicating extensive DNA scanning. They also examined phospho-mimetic and monoubiquitinated variants of D2-I, finding no significant differences from the unmodified complex, suggesting that these modifications do not impact DNA scanning.
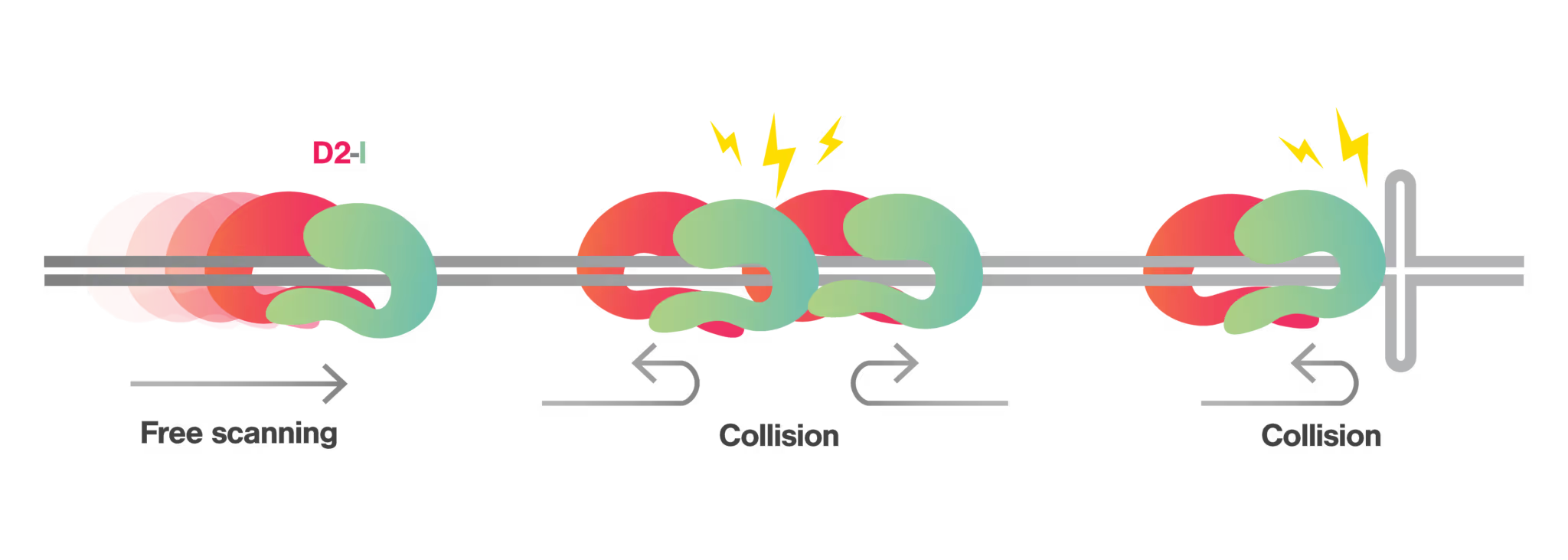
The researchers further analyzed how the D2-I complex reacts to obstacles on DNA. They introduced various roadblocks, such as the end of DNA, another D2-I complex, and a Holliday junction-like structure. Interestingly, rather than bypassing, detaching, or stalling at these obstacles, the D2-I complex typically altered its direction and continued sliding.
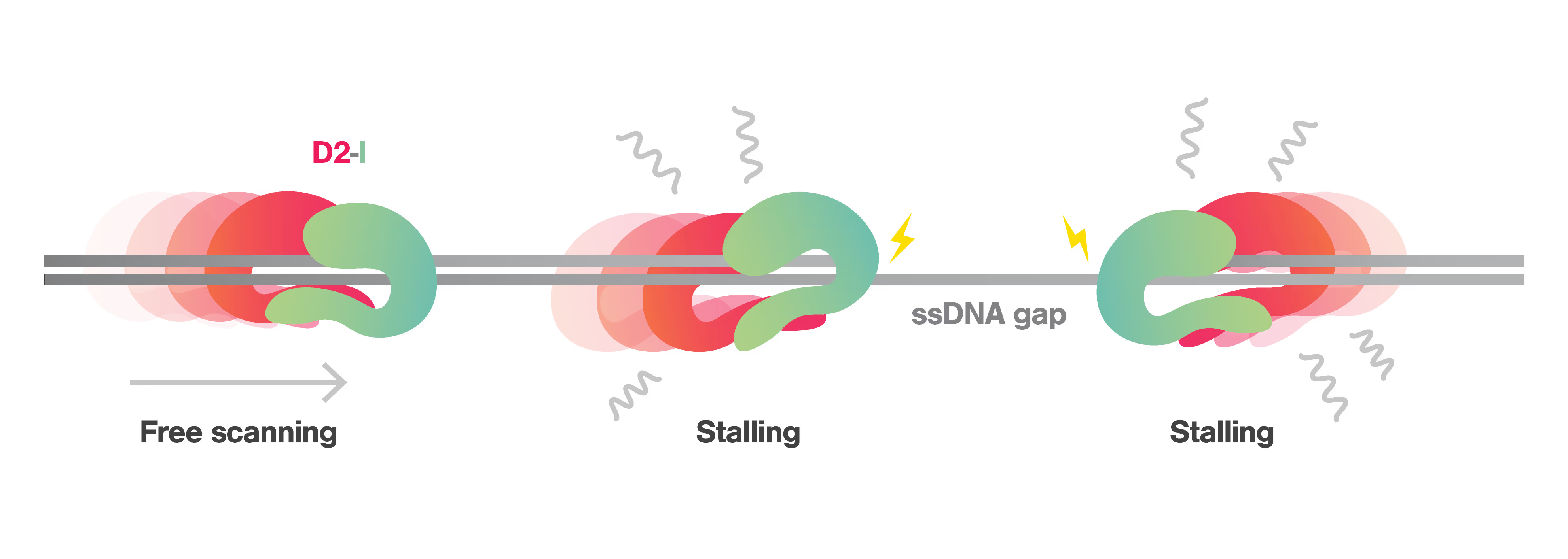
The behavior of the D2-I complex changes significantly upon encountering a double-strand and single-strand (ds-ss) DNA junction, a structure typical of replication forks crucial in DNA repair and replication processes. The D2-I complex was shown to slide on dsDNA but not on ssDNA. Notably, upon reaching a ds-ssDNA junction, it stalled for up to nearly one minute, highlighting a previously unknown role of D2-I in recognizing replication forks during crosslink repair and replication stress.
Lastly, the authors examined cryo-EM data of the D2-I complex clamping dsDNA and ss-dsDNA hybrids. The structure showed that dsDNA contacts the C-terminal domains of FANCI and FANCD2 proteins without specific binding positions, while ss-dsDNA junctions occupy a preferred binding position on D2-I. It confirmed the single-molecule data showing dsDNA sliding in the D2-I clamp, and D2-I stalling at ss-dsDNA junction. These data suggests that D2-I identifies and protects these junctions by clamping down at the junctions, thereby facilitating the initiation of DNA repair.
My lab has been studying the role of D2-I in DNA repair for many years. None of us had made the discovery that D2-I recognises a specific DNA structure. The C-trap was crucial for this as it allowed us to monitor the dynamics of D2-I on DNA. Understanding the dynamics was really the key to making progress.
Dr. Lori Passmore (MRC Laboratory of Molecular Biology), one of the corresponding authors of the publication
This study demonstrated that the D2-I complex can scan long, unobstructed double-stranded DNA, but stalls at single-strand-double-strand (ss-ds) DNA junctions. It revealed a previously unknown function of the D2-I complex in recognizing and protecting replication forks, which is only possible to resolve with the dynamic single-molecule technique. It’s the only technology that can monitor single protein’s translocation behavior on defined DNA substrates.
This new understanding of the D2-I complex marks a significant advancement in our comprehension of DNA repair processes. Not only does this breakthrough deepen our knowledge of the fundamental biology of the Fanconi anemia (FA) pathway but it also paves the way for new therapeutic approaches in treating cancer and other diseases linked to genomic instability. As research continues to decode the mysteries of DNA repair, the D2-I complex will undoubtedly remain a key focus in efforts to maintain genomic integrity.

FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions
DNA crosslinks block DNA replication and are repaired by the Fanconi Anemia pathway. The FANCD2-FANCI (D2-I) protein complex is central to this process as it initiates repair and is also known to play a more general role in DNA repair and in protecting stalled replication forks from unscheduled degradation.
Here, using single-molecule imaging, we investigate the behaivior of D2-I and provide a unified molecular mechanism that reconciles the roles of D2-I in recognition and protection of stalled replication forks in multiple DNA repair pathways.
Images adapted from the original research paper: Alcón, P., Kaczmarczyk, A.P., Ray, K.K. et al. FANCD2–FANCI surveys DNA and recognizes double- to single-stranded junctions. Nature (2024). https://doi.org/10.1038/s41586-024-07770-w