Discover the mechanisms and roles of chromatin organization and decipher the epigenetic code
Part of:
The dynamic architecture of DNA organization and chromatin remodeling
Quantify SMC activity, conformation and interactions at the molecular level
The direct accessibility of binding rates and localization of SMC complexes on DNA and the ability to quickly change buffer conditions delivers key insights into the molecular mechanism of interaction.

Case study
With this method, the labs of Johannes Stigler, Luis Aragon, and Shixin Liu were able to identify unique binding and mobility behavior of SMC5/6 on ss- vs ds-DNA and could identify DNA fork structures as preferred binding location, emphasizing SMCs’ critical role in DNA replication and repair (see figure).
The real-time single-molecule imaging capabilities of the C-Trap also enabled Frank Uhlmann and his collaborators to directly observe condensin interacting with multiple dsDNA molecules, a mechanism that had earlier been identified for cohesin using a similar C-Trap assay.
In summary, Dynamic Single-Molecule microscopy is key to quantifying
- Binding rates and location of SMC complexes on different DNA structures
- Dynamic movement of SMC complexes on DNA and looping/cross-linking activity
- Influence of additional factors (ATP, small molecules, other DNA-binding proteins) on SMC kinetics and dynamics
By applying tension on cohesin-looped DNA, Luis Aragon and collaborators were able to quantify loop size as well as mechanical stability of the cohesin-mediated cross-links.
Furthermore, tension can also be directly applied to SMC complexes (or subunits) to quantify their conformational dynamics under force. By doing so, Johannes Stigler and colleagues identified structures and intermediate states in the coiled-coil region of SMCs that are critical for dynamic folding of the complex. For more details on the assay, see our protein folding page.


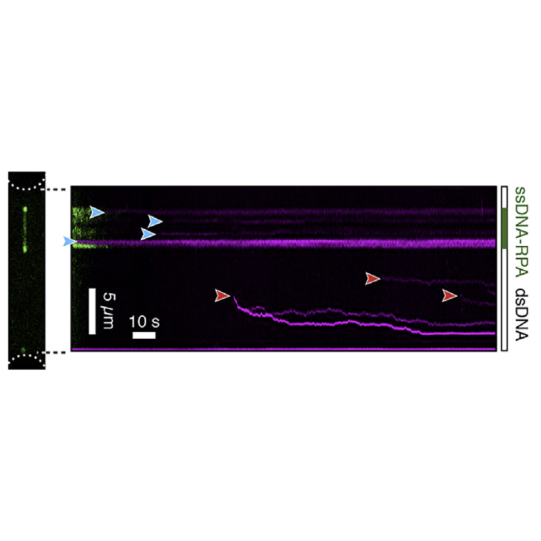
This kymograph shows SMC5/6 complexes binding to ssDNA-dsDNA hybrids. Complexes in ssDNA regions are static (blue arrowheads) while those in dsDNA regions are mobile (red arrowheads). Figures adapted from Tanasie et al. Cell reports (2022)

Force is applied to double-stranded DNA (dsDNA), exposing single-stranded DNA (ssDNA) around nick sites. RPA (green) selectively binds to those ssDNA regions. Smc5/6 (purple) binds to different regions of the DNA molecule (white arrowheads). This image shows 2D scan of the assay with ssDNA, dsDNA, RPA (green) and SMC5/6 (purple) bound on ssDNA.
Explore further

Insights into Bacterial DNA Condensation and chromosome organization
Guided by our host, a renowned expert in bacterial cell biology, Dr. Fernando Moreno Herrero from the Department of Macromolecular Structures, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas, Madrid, we will explore the critical ParABS partitioning system and the SMC complex, which serve as the driving forces behind bacterial chromosome segregation.
Using cutting-edge single-molecule techniques and optical tweezers combined with a confocal microscope, we offer an exclusive view into how cytidine triphosphate (CTP) binding and hydrolysis play integral roles in the interaction between parS and ParB.
Follow chromatin remodeler activity in real-time
The single-molecule fluorescence imaging module of the C-Trap is designed to follow the dynamic movement of chromatin remodelers on DNA, also in the presence of nucleosomes or other structuring elements.
Multi-color imaging enables the tracking of different species of proteins involved in the process and gives access to quantitative, real-time information on:
- Binding kinetics and location of chromatin remodelers, histone chaperones, and other components
- Dynamic behavior of nucleosome remodelers on DNA, e.g. sliding and diffusion
- Colocalization of remodelers with nucleosomes, nucleosome transport, histone eviction

Case study
The groups of Taekjip Ha, Carl Wu and Shixin Liu have extensively used LUMICKS’ C-Trap to unravel details of remodeler target search and to discover hitherto unknown nucleosome remodeling capabilities of origin recognition complexes (ORCs), respectively.
The optical traps of the C-Trap allow the application of well-defined mechanical tension on a single DNA molecule, mimicking the case of incomplete sister chromatid segregation or other mechanically challenging scenarios DNA is regularly facing.
By doing so, the groups of Erwin Peterman and Gijs Wuite in collaboration with Ian Hickson discovered an unknown role of the checkpoint helicase PICH: On DNA under tension, PICH shows nucleosome unwrapping and histone transporting activities, as directly observed with correlative dynamic single-molecule microscopy (Figure 1).




Kymograph showing PICH-eGFP (green) and Atto647N labelled histones on DNA under tension (12.5 pN). A PICH-eGFP dimer initiates histone sliding at the instant of collision (white arrow) with the Atto-647N labelled histones. Top: PICH channel (green) Center: Histone channel (red) Bottom: merge Figure adapted from Spakman et al. Nature Communications (2022)
Use the force: Quantify nucleosome stability and cross-linking
By tethering arrays of nucleosomes between two optical traps, you gain access to quantitative information about the mechanical stability of individual nucleosomes as well as possible cross-linking interactions.
Additional factors like histone-modifying enzymes or nucleosome cross-linkers are quickly added via the integrated microfluidics and their interaction with the DNA/nucleosomes can be observed in real-time with single-molecule fluorescence microscopy.
The effects these factors have on nucleosome stability and cross-linking can be directly assessed by increasing the tension on the DNA substrate (see figure): Depending on the modification, the accessibility of DNA can be increased (destabilization, lower force required) or decreased (stabilization, higher force required).
In summary, with the dynamic single-molecule (DSM) approach you gain quantitative access to the
- Location, duration and sequential order of modifier-nucleosome interactions
- Mechanical stability of native and modified nucleosomes
- Strength of nucleosome-nucleosome interactions mediated by cross-linking factors

Case study
Amongst other labs, the groups of Mark Williams and Shixin Liu used this method successfully to unravel the impact of histone chaperones on nucleosome (dis)assembly, and Polycomb complex-mediated nucleosome cross-linking, respectively.




Top: Schematic representation of the molecular construct: a series of 12 3 207-bp Widom 601 positioning sequences flanked by two tagged handles, each labeled by digoxygenin/biotin tags for bead attachment Bottom: Cycles of array extension/release disrupt nucleosomes in distinct stages that are only partially reversible upon release. Polymer models (solid lines) bracket outer wrap release (red), a single inner wrap disruption (blue), and the final full DNA (magenta). Shaded regions denote work done during unwrapping. Release partially restores wrapping. Figure adapted from McCauley et al. Cell Reports (2022)
C-Trap
Biomolecular interactions re-imagined
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
Direct visualization of the effect of DNA structure and ionic conditions on HU–DNA interactions
A conserved ATP- and Scc2/4-dependent activity for cohesin in tethering DNA molecules
Reversible histone glycation is associated with disease-related changes in chromatin architecture
SITC 2025
CAR-TCR Summit 2025
CICON 2025
























































































