In a recent article published in Nature Communications, Sanchez et al. successfully employed LUMICKS’ C-Trap® to discover new insights into the molecular dynamics of initial replisome formation. In a collaborative effort between Prof. Dekker (TU Delft) and Prof. Diffley (Francis Crick Institute) laboratories, the scientists uncovered that origin recognition complex (ORC) is a mobile protein that can scan the DNA for its target, and established the existence of a dynamic, single-hexameric form of the Mcm2-7 (MCM) helicase.
The first step of replisome assembly initiates by loading of several proteins to the DNA including the double-heteromeric MCM helicase. Despite the extensive knowledge about the individual components of the replisome formation, little is known about the dynamics of the individual proteins and their contribution to the complex formation. Thus, here employed single-molecule techniques provide unprecedented information expanding our understanding of the replication machinery assembly.
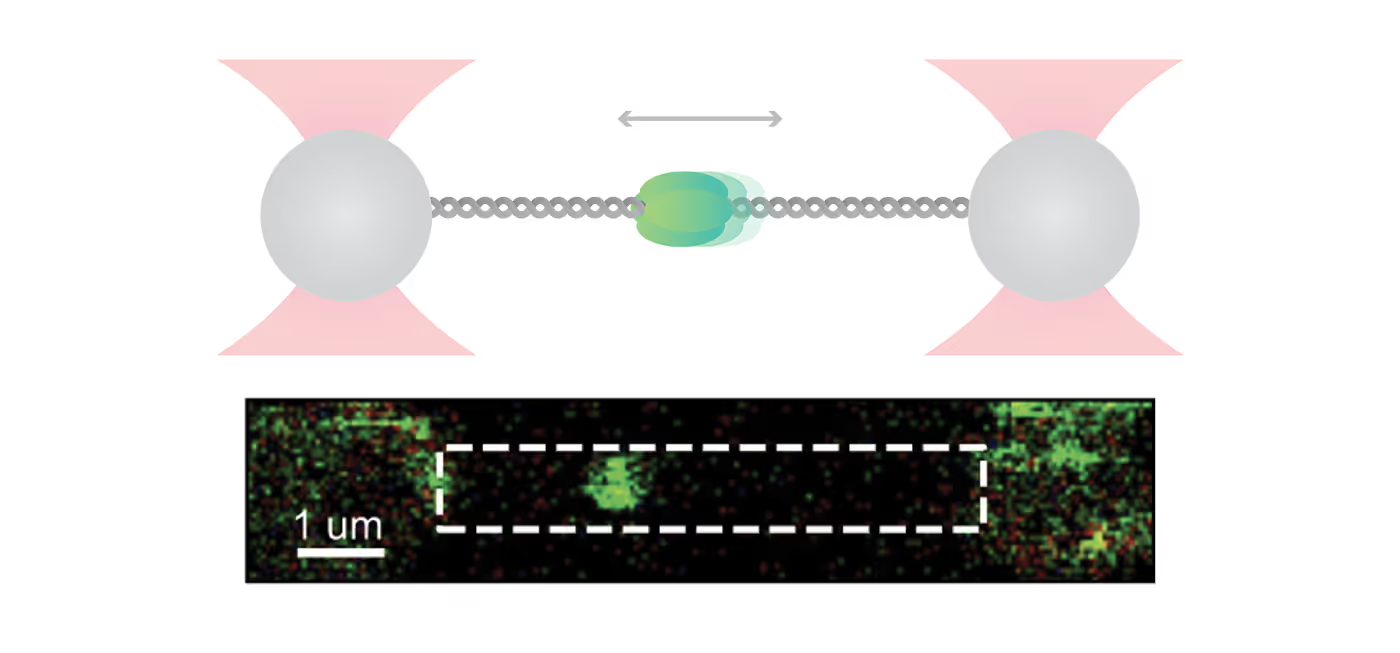
For their investigation, the researchers used C-Trap® optical tweezers correlated with a multicolor confocal microscopy system to hold a DNA strand and directly visualize the binding dynamics and motion of fluorescently labeled ORC and MCM proteins. The incorporated microfluidics of the system allowed for an easy step-by-step assembly of individual proteins on DNA.
In their experiments, the scientists discovered that ORC is a mobile protein that performs a linear target search for origin. Initially, ORC binds non-specifically to DNA and subsequently diffuses until it finds the origin sequence, where it halts its movement. This reduction in mobility might allow MCM recruitment in the preferred location.
Beyond that, the researchers uncovered that next to the known double-hexameric (DH) form, the recruited MCM helicase was also present in a dynamic, single-hexameric (SH) form. The authors suggest that DH might be formed by the encounter of two SH MCMs upon diffusion and that the current “MCM paradox” – recruitment of many more MCM than are actually used – may be connected to the large population of SH forms that was found in this study.
“Our results demonstrate that effective helicase loading relies on an interplay between protein diffusion and origin recognition, and suggest that MCM is stably loaded onto DNA in multiple forms”, the authors conclude.
Read the full article titled “DNA replication origins retain mobile licensing proteins” that is published in Nature Communications.