In two recent scientific publications, Johannes Stigler and his team from the Gene Center of the Ludwig-Maximilians-University Munich, Germany, set out to mechanistically describe the action of the Structural Maintenance of Chromosome (SMC) Complexes using LUMICKS’ C-Trap® technology. By performing dynamic single-molecule experiments coupled with real-time visualization, they were able to elucidate the mechanisms governing SMC-DNA interactions (Tanasie et al, Cell Reports 2022). Notably, in what constitutes a major milestone for LUMICKS as it is the first study to use C-Trap data to study protein folding, they were also able to uncover conformational features that confer the SMC Complex its function (Freitag et al, Biophysical Journal 2022).
SMC complexes are essential for chromosome maintenance and organization, as they ensure efficient genome replication and segregation and are in charge of DNA repair. SMC proteins consist of long anti-parallel coiled coils (CCs), a hetero-dimerization domain and an ATP-binding domain at opposing ends.
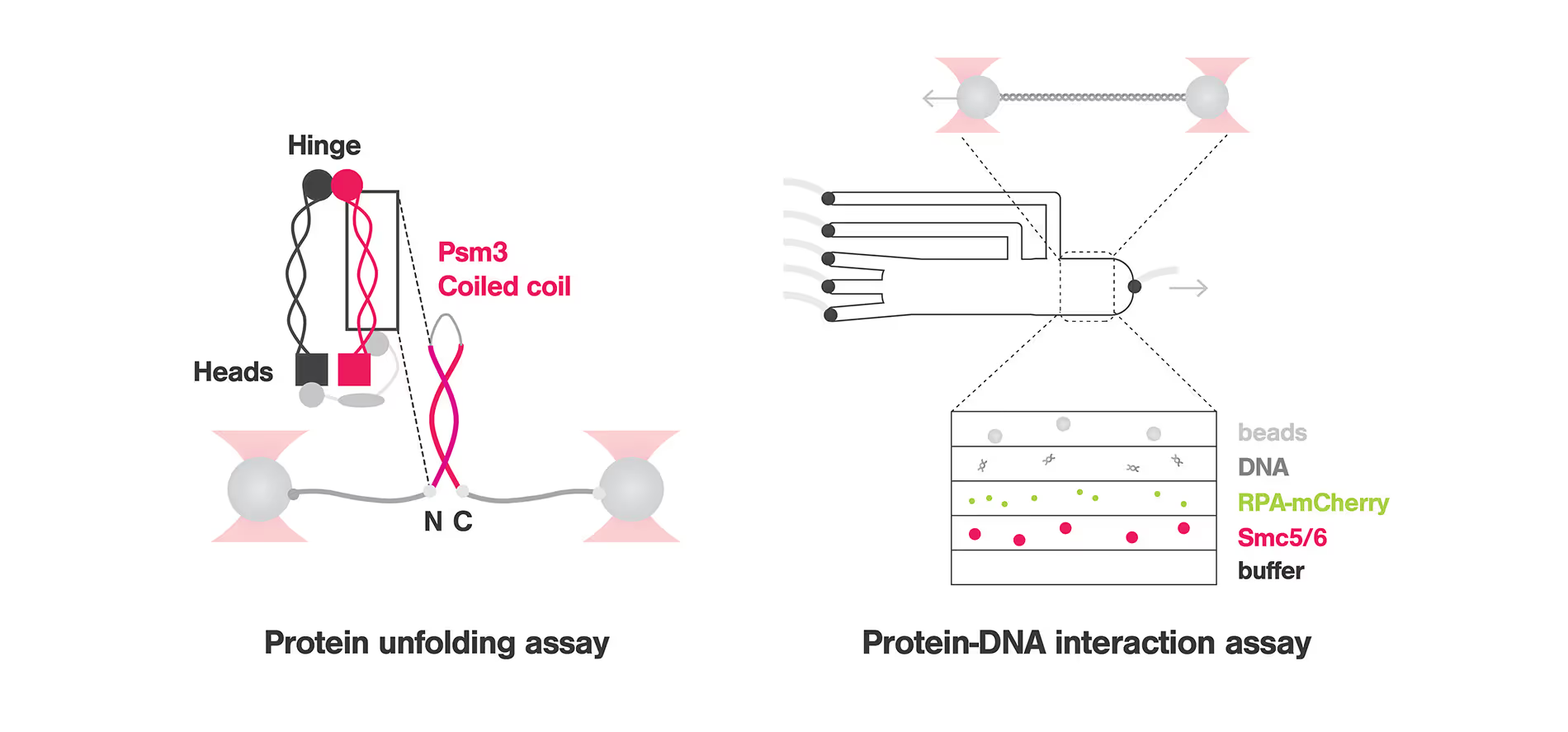
To understand the mechanism by which SMC Complexes are recruited to and interact with DNA, Nicoleta-Loredana Tanasie and colleagues studied Smc5/6, a member of the SMC family that has been linked to replication fork-related processes to understand how this protein is recruited to DNA lesions. Using the C-Trap that combines optical tweezers, confocal microscopy and microfluidics with multiple laminar flow channels, the researchers used a hybrid ssDNA-dsDNA that mimicked the replication fork and fluorescently labelled the ssDNA for visualization. These experiments highlighted that Smc5/6 is enriched to ssDNA and ssDNA-dsDNA junctions while it is depleted at dsDNA fragments. Further tests proved that Smc5/6 scans the dsDNA in a 1D diffusion fashion and stops when it reaches the fork junction. Moreover, varying the distance between the trapped beads to create tension on the DNA filaments, confirms that Smc5/6 stabilizes ssDNA-dsDNA fork junctions.
To study the structure-function relationship of the SMC Complex, Marvin Freitag and colleagues investigated how the conformational changes of the CC domain affect the protein function of Psm3. Unfolding experiments were carried out using the C-trap, in which a large single-peptide piece of the CC domain was attached to DNA handles and tethered between optically trapped beads. These experiments revealed that Psm3 has a general zipper-like unfolding pattern that goes through 3 intermediates: a shutter, the elbow and a non-predicted feature. Importantly, replacing the elbow leads to misfolding, pointing to the function of the elbow as a folding guide that ensures the proper arrangement of the protein and therefore its function. This discovery could help in the development of new therapies targeting SMC proteins and related diseases.
As the authors say: “Optical tweezers allow the study of protein folding features with high temporal and spatial resolution on a single molecule basis.”
For further information about these studies, check out the papers: “Stabilization of DNA fork junctions by Smc5/6 complexes revealed by single-molecule imaging” and “Single-molecule experiments reveal the elbow as an essential folding guide in SMC coiled-coil arms,” published, respectively, in Cell Reports and Biophysical Journal.
Are you interested in using dynamic single-molecule tools like the C-Trap for your research? Do not hesitate to contact us for more information, a demo, or a quote.
Top image adapted from Tanasie et al, Cell Reports and Freitag et al, Biophysical Journal, 2022.