Redefine nucleic acids research and observe every conformational change at the single-molecule level
- Observe individual RNA or DNA molecules transitioning in real time among folded, misfolded, and intermediate conformations
- Reveal how proteins, ligands, drugs or sequence elements reshape nucleic acid folding and structure landscapes
- Quantify structural, kinetic, and energy landscape parameters—even for highly complex RNA molecules
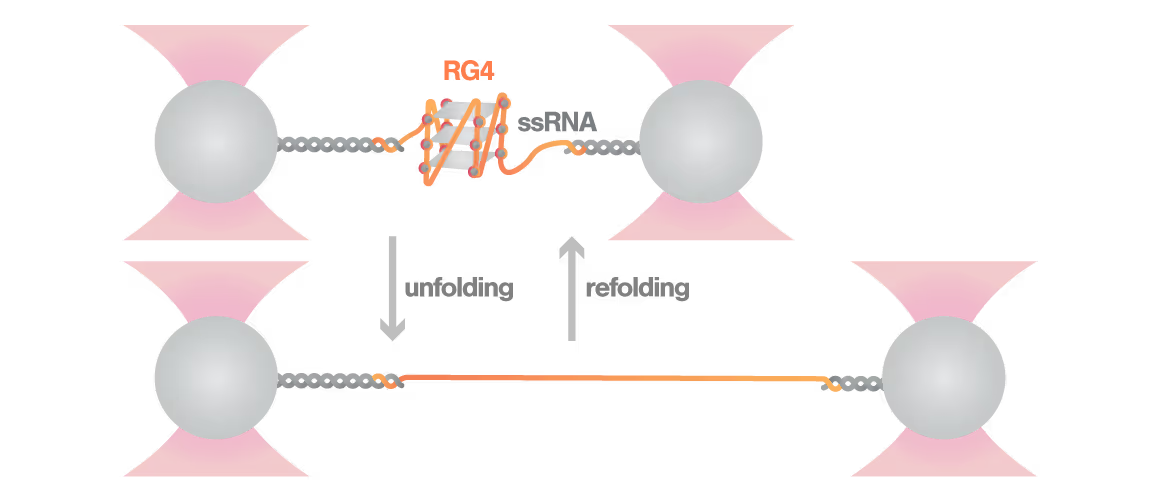
Revealing stability and dynamics of telomeric G-quadruplexes
G-quadruplexes are specialized DNA and RNA structures that form in guanine-rich regions critical for telomere integrity and cellular aging. These dynamic structures regulate gene expression and chromosome stability, with disruption implicated in cancer development and aging-related diseases. Despite extensive study, scientists cannot directly observe how G-quadruplexes fold and function in real biological contexts—limiting our ability to target them therapeutically. With cancer affecting millions worldwide, understanding these structures has become essential for developing novel targeted treatments.
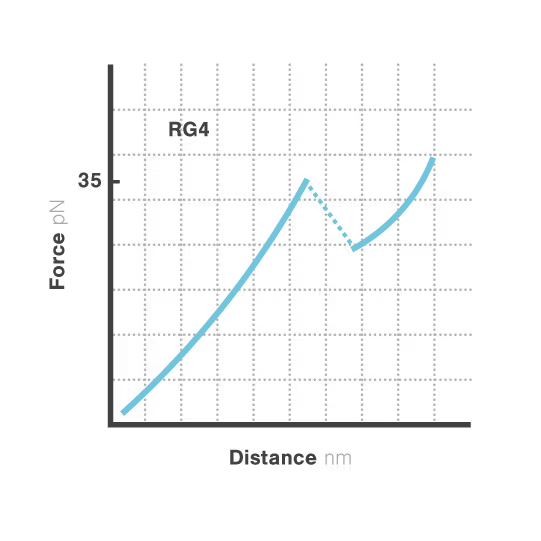
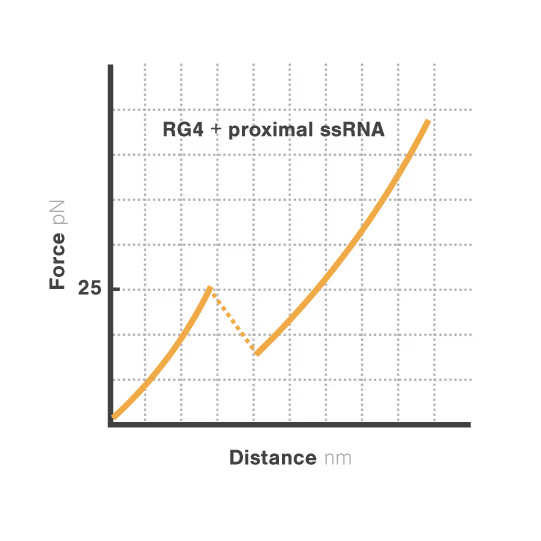
A study led by Bo Sun at ShanghaiTech University explored how human telomerase RNA (hTR) forms and maintains stable G-quadruplex structures. Using LUMICKS C-Trap, they monitored real-time folding and unfolding of single hTR molecules, revealing:
- Multiple distinct G-quadruplex conformations exist, identifiable only by single-molecule force spectroscopy.
- Local RNA sequences strongly modulate the unfolding and refolding kinetics of these critical telomeric structures.
- These novel insights into G4 structural dynamics offer novel therapeutic strategies for targeting telomere-related diseases, including cancers and aging.
Dive into the publication
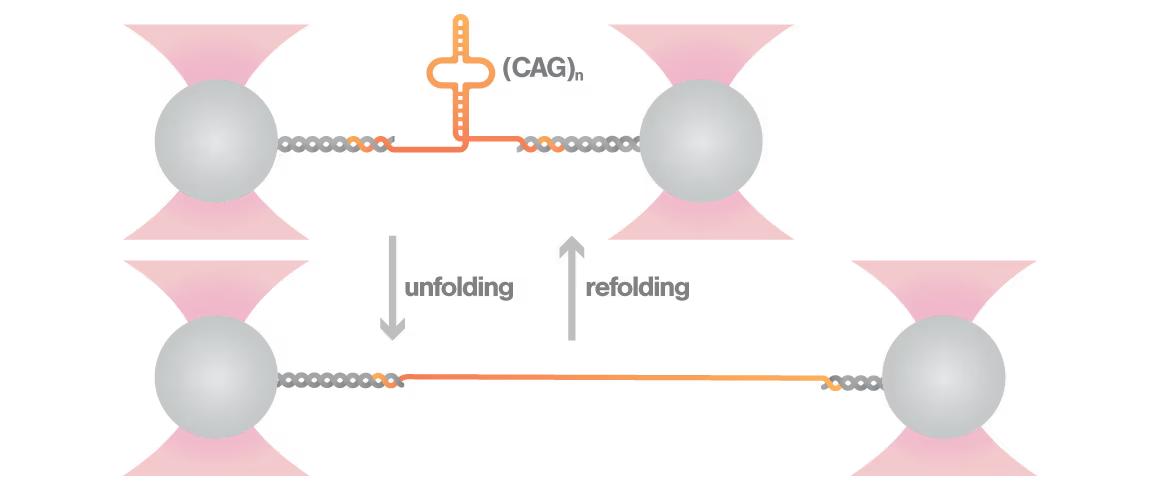
Elucidating toxic RNA misfolding dynamics in Huntington’s disease
Expanded repeat sequences in RNA form abnormal structures linked to numerous neurodegenerative diseases including Huntington's, ALS, and certain ataxias. These toxic RNA structures aggregate in cells and disrupt normal neuronal function, triggering progressive neurodegeneration. Despite significant advances in genetic diagnosis, researchers cannot visualize how these repeat expansions actually misfold and aggregate —hampering therapeutic development. With neurodegenerative disorders affecting over 50 million people globally, understanding RNA structural dynamics has become critical for developing effective treatments.
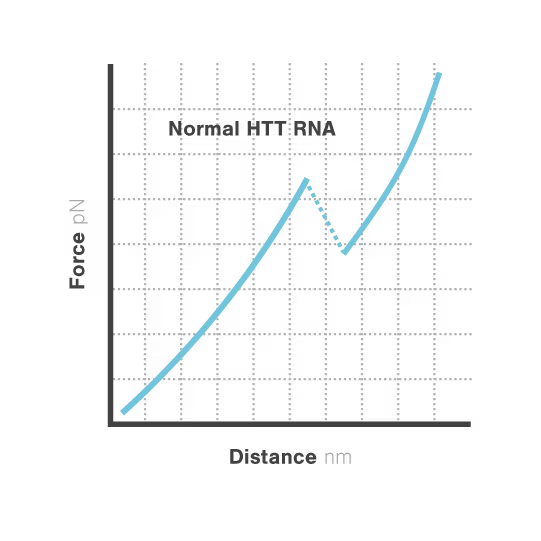
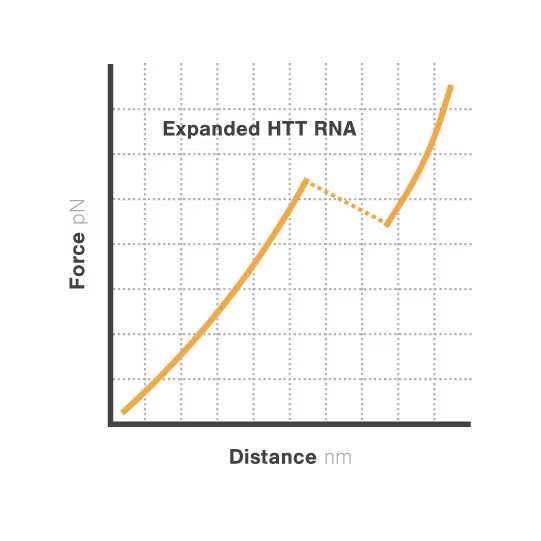
Researchers Christian Kaiser and Sarah Woodson investigated how expanded CAG repeats in HTT RNA influence misfolding and aggregation. By applying real-time single-molecule force measurements with LUMICKS C-Trap, they discovered:
- Healthy (normal) HTT RNA unfolds cooperatively in a single well-defined step.
- Expanded repeats unfold stepwise (“stick-slip” pattern) at lower forces, indicating non-cooperative behavior.
- Frequent partially unfolded structures self-associate, directly linking RNA misfolding dynamics to Huntington disease progression
Dive into the publication
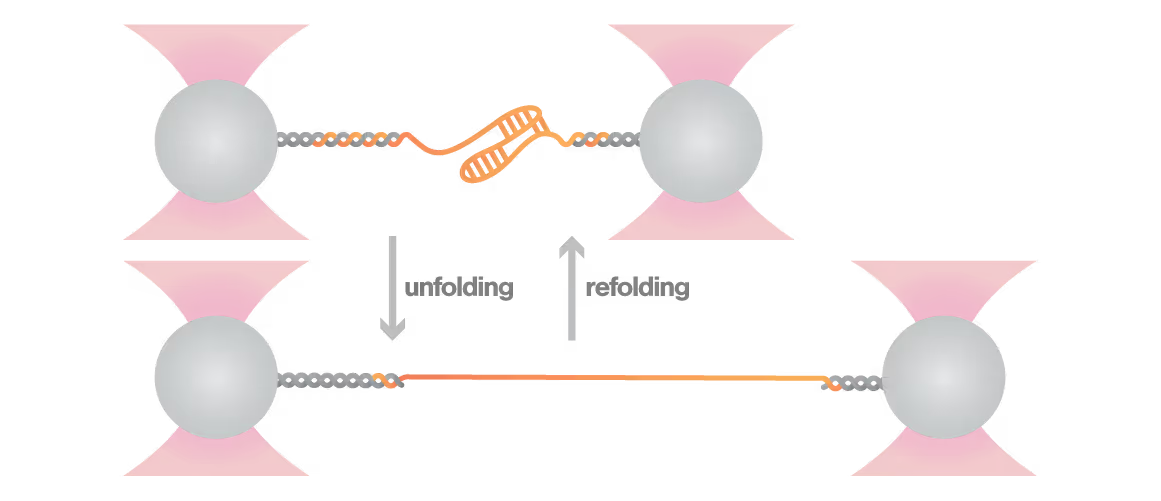
Protein-mediated frameshift regulation in SARS-CoV-2
RNA pseudoknots are complex folded structures that regulate critical processes in viral replication and gene expression. These dynamic elements enable viruses to produce multiple proteins from limited genetic material and evade host defenses through conformational changes. Despite their central role in viral pathogenesis, scientists cannot directly observe how these structures interact with host proteins during infection—limiting antiviral development. With viral diseases continuously threatening global health security, understanding RNA structural dynamics has become essential for creating next-generation antivirals.
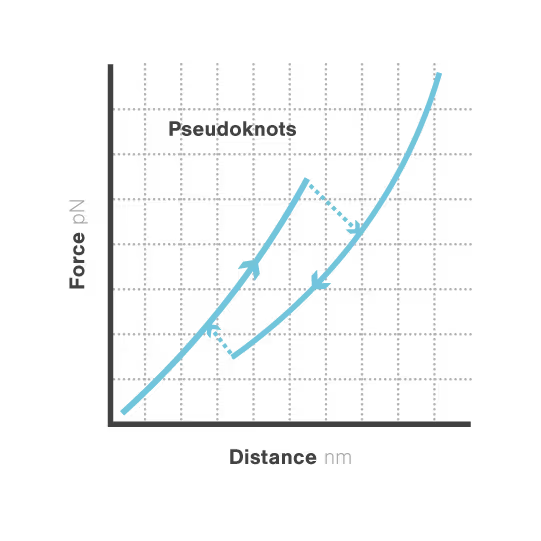
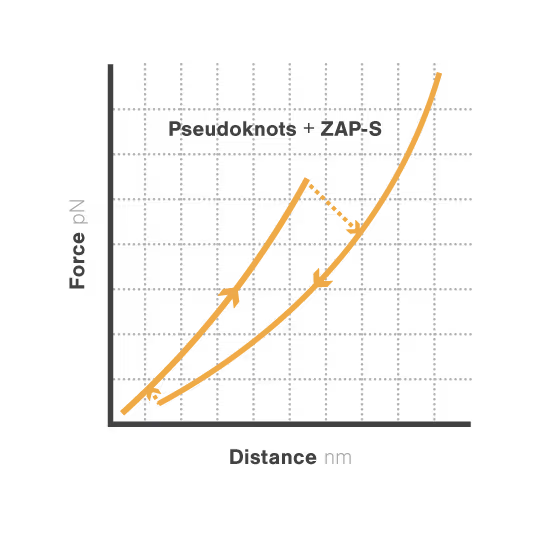
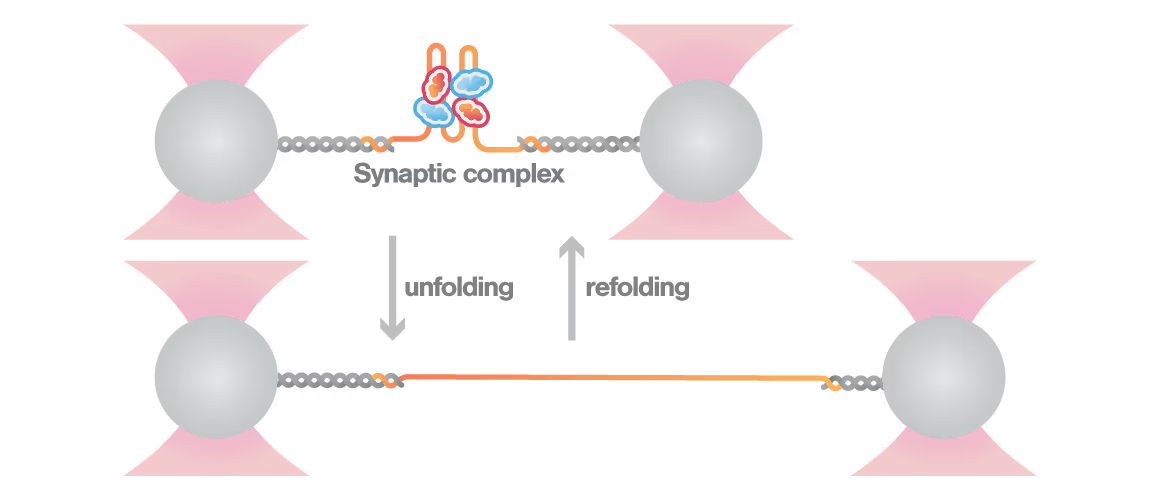
A study led by the Neva Caliskan Lab at the Helmholtz Institute for RNA-Based Infection Research investigated how ZAP-S protein modulates SARS-CoV-2 frameshifting through altering the pseudoknot structure. By directly tracking the real-time pseudoknot structural dynamics in the presence or absence of ZAP-S using LUMICKS C-Trap, they revealed:
- ZAP-S binding destabilizes the RNA pseudoknot structure, preventing its refolding and impairing frameshifting activity essential for viral protein production.
- Reduced frameshifting efficiency means fewer essential viral proteins are produced, weakening the virus’s ability to replicate.
- Ultimately, viral replication is significantly reduced, highlighting ZAP-S interaction with the pseudoknot as a novel, targetable antiviral mechanism with clear translational potential.
Dive into the publication
A novel targetable mechanism against antibiotic resistance: the structural stability of bacterial integrons
Bacterial integrons are genetic elements that enable bacteria to capture external genes and develop antibiotic resistance. These molecular machines incorporate resistance genes through DNA recombination, driving the spread of untreatable infections. Despite decades of research, scientists still cannot explain why some genes transfer efficiently while others don't—a critical gap hindering our ability to predict resistance spread. With resistant infections causing over 1.2 million deaths annually, understanding these mechanisms has become urgent.
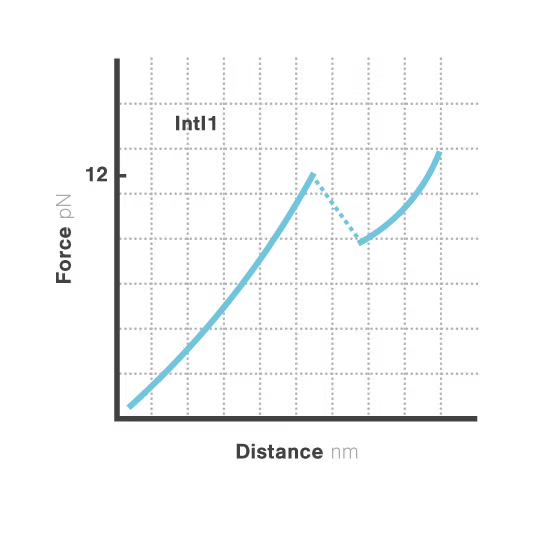
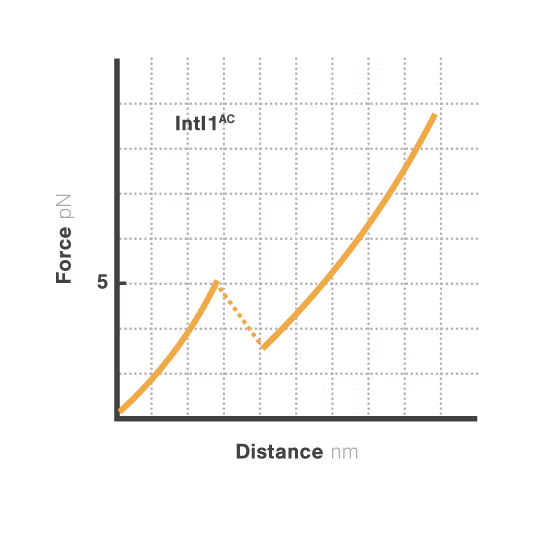
A breakthrough study led by Michael Schlierf at TU Dresden in collaboration with Didier Mazel at Institut Pasteur investigated this mystery using single-molecule techniques. By applying controlled forces to individual DNA-IntI1 integrase complexes with LUMICKS C-Trap, they revealed:
- Efficiently recombining DNA sites form mechanically stable complexes with the IntI1 protein that resist forces up to high forces.
- Poorly recombining DNA sites form weaker protein-DNA complexes that break apart at lower forces.
- The mechanical stability of these IntI1-DNA complexes directly correlates with recombination efficiency in living bacteria, revealing a physical basis for genetic adaptation and a potential target for combating antibiotic resistance.
Dive into the publication
C-Trap
Molecular biology as never seen before
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
A Minimal Load-and-Lock Ru Luminescent DNA Probe
The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting
Optical Tweezers to Study RNA-Protein Interactions in Translation Regulatio
SITC 2025
CAR-TCR Summit 2025
CICON 2025