The intricate mechanistic details of repair proteins
- Observing repair processes in real-time
- Studying induced structural changes
- Extracting high-resolution functional information
Revealing molecular mechanism heterogeneity in UV-DDB-related DNA repair processes
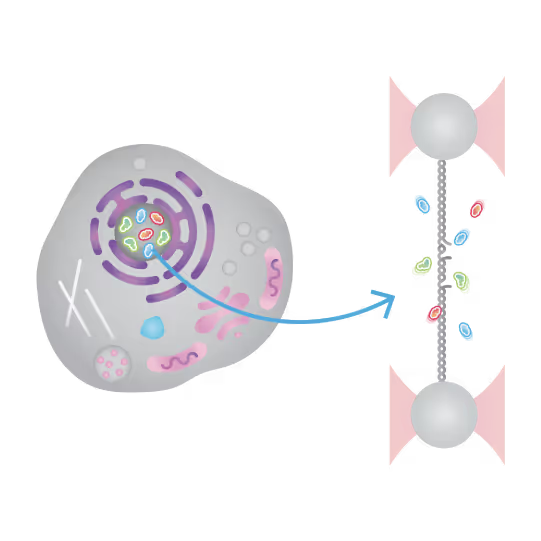
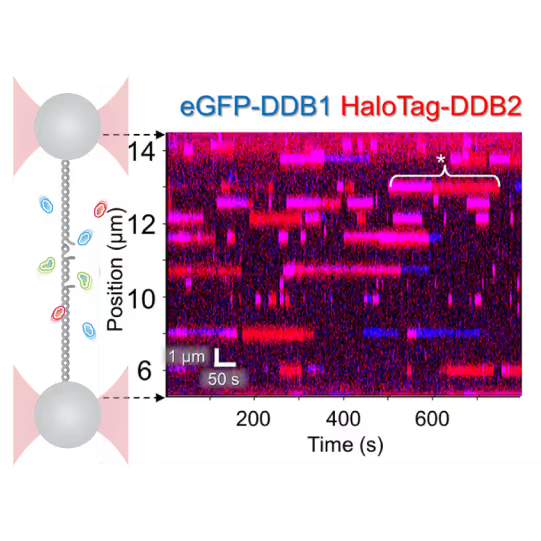
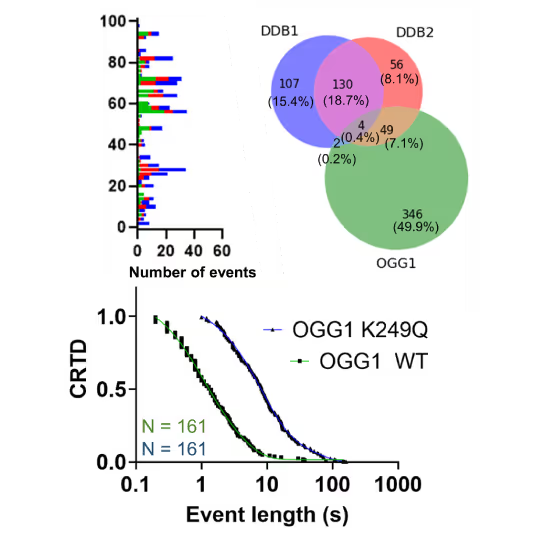
Understanding the sequence of events in assembling multi-protein complexes is crucial for comprehending DNA repair processes and identifying potential drug targets. LUMICKS’ dynamic single-molecule approach provides such insights, even within the physiological environment of cellular extracts.
Professor Ben Van Houten‘s team at the University of Pittsburgh successfully used cellular extracts without complex protein purification to:
- Track multi-step binding processes
- Quantify binding kinetics
- Reveal molecular mechanism heterogeneity in UV-DDB-related DNA repair processes
Furthermore, using the C-Trap’s ability to apply precise mechanical stress on individual molecules, they discovered a correlation between PARP1 binding kinetics to nicks, and the mechanical state of the DNA substrate.
Dive into the publication

Single-molecule analysis of cancer DNA-protein interactions from nuclear extracts
Understanding of DNA repair mechanisms could advance treatments for cancer and diseases of aging. But reconstituting DNA repair protein complexes from cancerous tissues to study their mechanisms of action is often time-consuming or, in some cases, impossible. A new technique performing dynamic single-molecule analysis directly on nuclear extracts allows rapid mechanistic analysis of mutant proteins from cancer cells, providing previously unseen insights into their mechanisms of action. This new innovative tool, when combined with rapid data analysis, represents a bridge between the study of biochemistry of purified proteins and molecular biology.
Visualizing DNA translocation and lesion recognition
Static structural data often limits hypotheses and models explaining DNA-protein binding mechanisms, failing to capture complex dynamics.
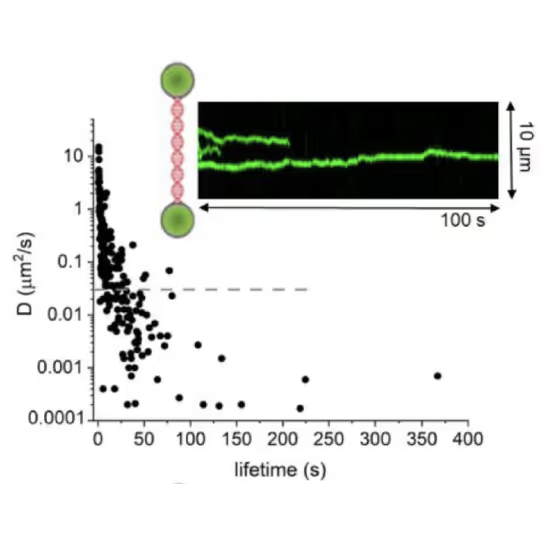
Ingrid Tessmer (Rudolf Virchow Centre Würzburg) and her team employed the C-Trap to directly visualize DNA translocation and lesion recognition by O6-alkylguanine DNA alkyl-transferase (AGT).
This approach unveiled details of AGT’s capabilities to:
- Dynamically bind DNA
- Form clusters
- Search lesions
Real-time observation with the C-Trap provided quantitative insights, challenging established models of AGT’s unidirectional movement, previously believed to be accelerated by cluster formation.
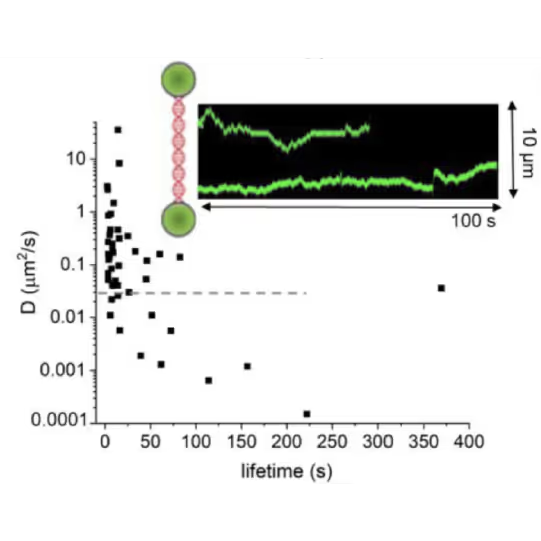
DNA lesion search dynamics. One-dimensional diffusion constants (D) on DNA plotted over the lifetimes of complexes on the DNA for AGT. The insets show representative kymographs (green traces) obtained by fluorescence microscopy-coupled dual trap optical tweezers, in which the y direction corresponds to the positions on the DNA tether (shown schematically between two beads held in the two optical traps), and the x direction to time.
Dive into the publication

A deep dive into Nucleotide Excision Repair (NER) and its crucial role in Alkyl-DNA lesion repair and cancer prevention
In this session, hosted by DNA repair expert Dr. Ingrid Tessmer, Rudolf Virchow Center for Experimental Biomedicine, University of Würzburg, we dive deep into the role of alkyltransferase-like proteins (ATLs) and their role in NER. Despite their inherent catalytic inactivity, ATLs play a remarkable role in targeting alkyl lesions for repair by the NER system. Through a combination of single-molecule and ensemble methodologies, a detailed view of the recruitment process of UvrA – the initiating enzyme of prokaryotic NER – to an alkyl lesion by ATL has been observed for the first time.
Moreover, we delve into the mechanisms of lesion recognition by ATL, and illustrate the dynamic DNA lesion search undertaken by highly active ATL and ATL-UvrA complexes.
Don’t miss this opportunity to broaden your understanding of DNA repair and its potential role in revolutionizing cancer treatment strategies.
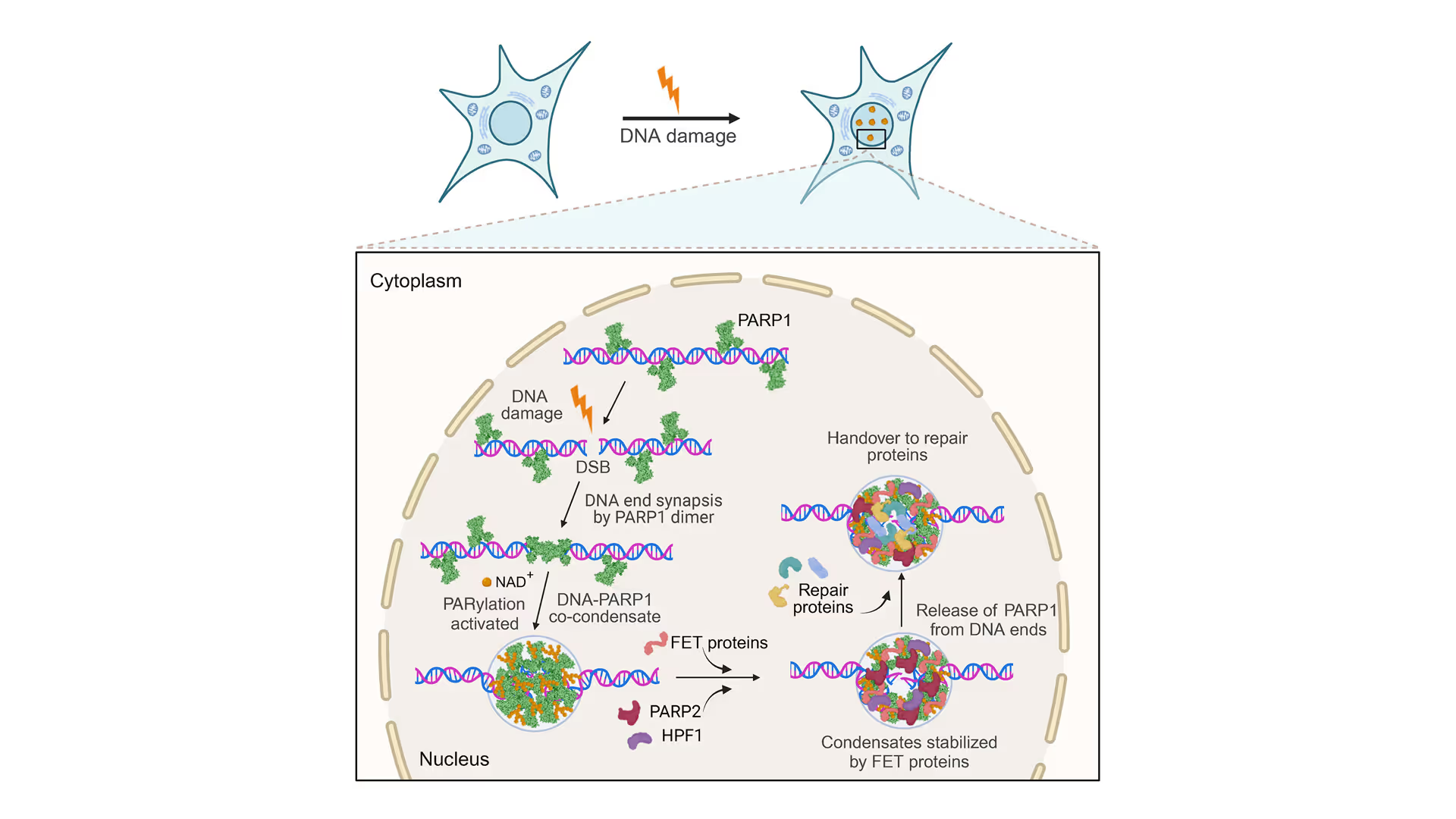
Revealing the facilitation of DNA repair through PARP1 condensation
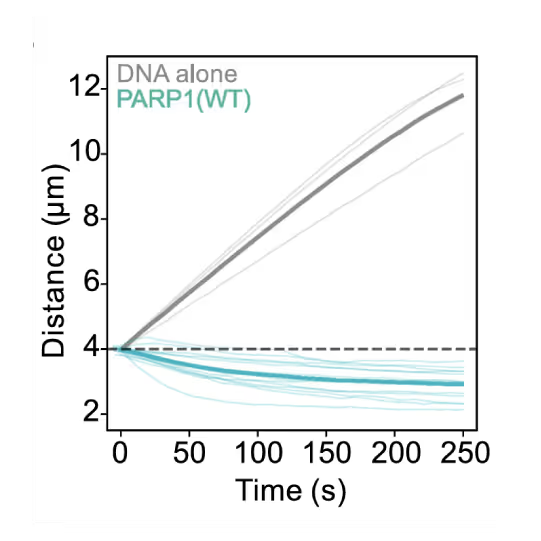
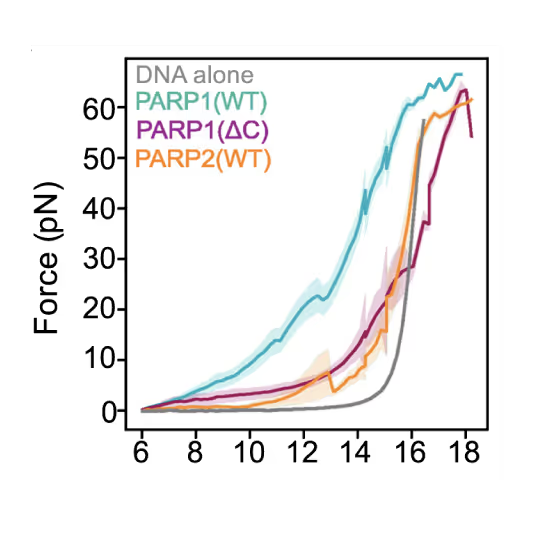
Preventing DNA end disjunction is vital for DNA double-strand break (DSB) repair, yet the role of PARP1 in this process remains unclear.
Simon Alberti (MPI-CBG Dresden) and collaborators used LUMICKS’ dynamic single-molecule approach with the C-Trap to investigate PARP1 recruitment to DSBs. Single-molecule imaging, combined with measuring mechanical forces, revealed that:
- PARP1 forms condensates around damage sites
- These biomolecular condensates physically connect DNA ends, even under tension
- Condensate formation facilitates the recruitment of additional repair and regulatory factors
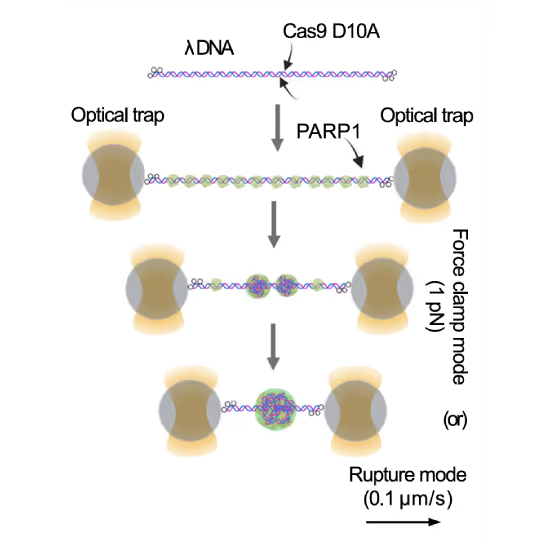
Representative florescence images of mEGFP-tagged PARP1(WT), PARP1(ΔC), and PARP2(WT) localization on tethered λ DNA, which was nicked on both strands by CAS9(D10A) with a 60 bp gap between the nicks. Only PARP1(WT) shows condensation at the damage site. Scale bar, 2 μm. Image source: Chappidi et al. Cell 2024
Dive into the publication
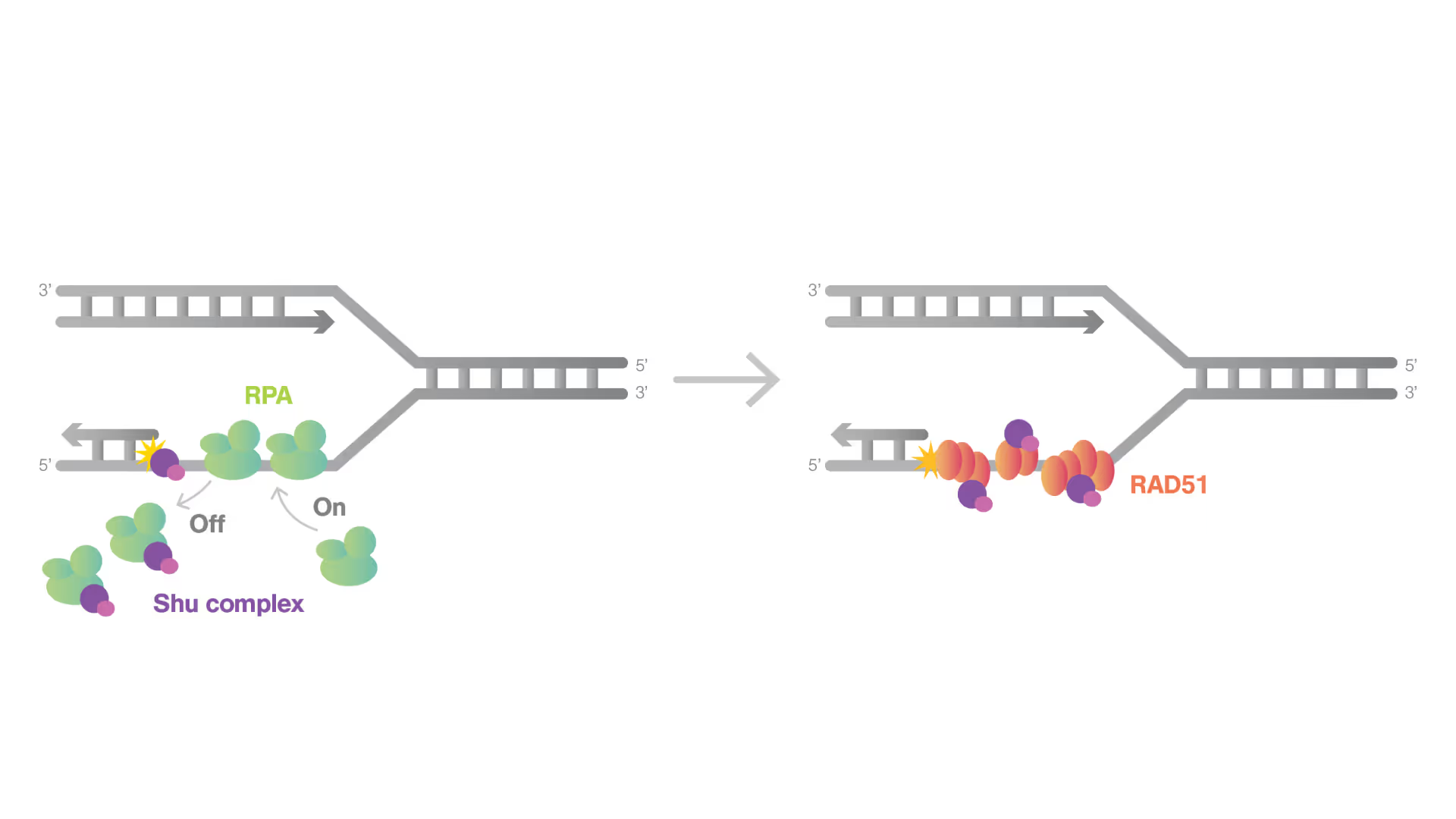
Dynamic Single-Molecule revealed molecular mechanism of homologous recombination (HR) in DNA repair
Homologous recombination (HR) is crucial for repairing double-strand breaks (DSBs) in DNA, yet some molecular mechanisms and protein roles remain unclear.
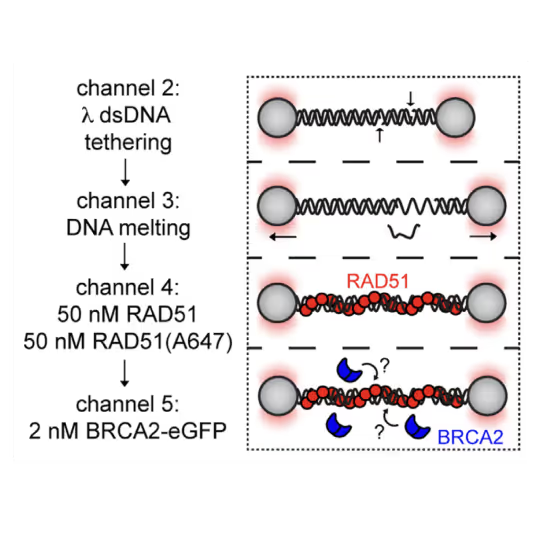
Utilizing the C-Trap, studies led by Steve West, Simon Boulton (both at The Francis Crick Institute, London), and David Rueda (Imperial College, London) uncovered new sub-steps and regulatory functions in RAD51 filament formation, a key HR process:
- Real-time observations revealed BRCA2’s role in nucleating and stabilizing RAD51 on RPA-coated ssDNA
- A diffusion-assisted mechanism involving dsDNA binding and sliding was identified
- Additionally, RFS-1/RIP-1 was found to act as a dynamic ‘chaperone’ to promote filament growth
- BCDX2 in turn was found to stimulate RAD51 filament nucleation and extension, dependent on RAD51B and RAD51C ATPase activities.
These findings elucidate two distinct BRCA2-dependent RAD51 loading mechanisms onto ssDNA and illuminate the roles of recombination mediators during filament growth.
Understanding these processes at the single-molecule level clarifies the function of BCDX2 and other factors in RAD51 assembly on ssDNA, which is crucial for replication fork protection and DSB repair, and essential for tumor avoidance.

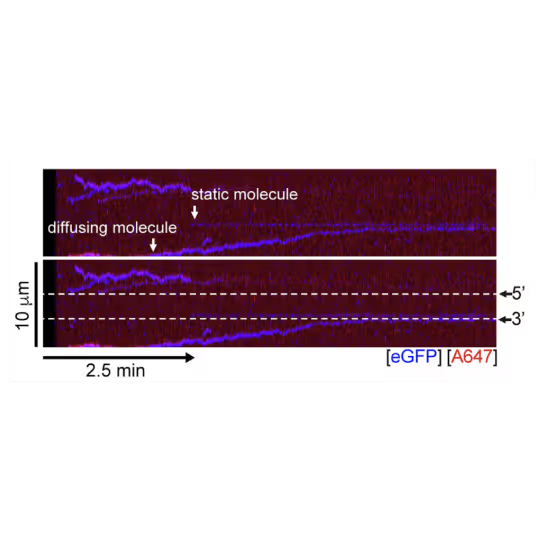
A representative kymograph showing diffusion-driven delivery of BRCA2-RAD51 complexes to ssDNA in the vicinity of the ds-ssDNA junction. Static BRCA2-eGFP molecules bound directly to the ssDNA gap. Mobile BRCA2-eGFP molecules diffuse along dsDNA. 25 nM RAD51-A466 (red) was incubated with DNA in the presence of 5 nM BRCA2-eGFP (blue) and 1.25 nM RPA in the presence of 5 nM SYTOX Orange. Position of the ssDNA gap is indicated by dashed lines.
Dive into the publication
Explore further

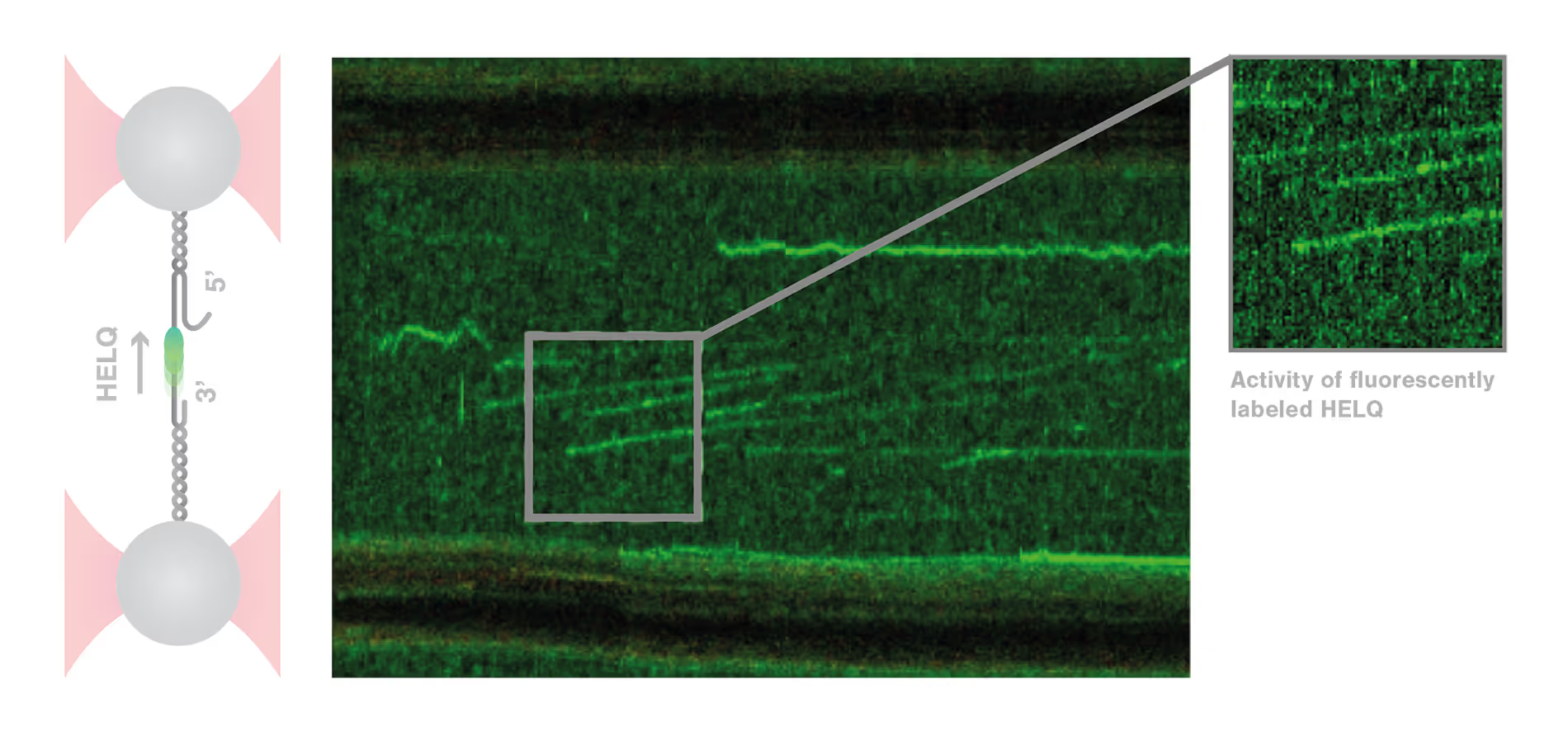
Dynamic single-molecule analysis offers invaluable insights into DNA repair mechanisms: HELQ in DSB case study
DNA repair is a highly complex and dynamic process that involves the interplay of numerous different proteins and components. The helicase HELQ is known to play a role in double-stranded breaks (DSBs) repair, but its molecular mechanisms remain unknown.
A study by the research group led by Simon Boulton presented how dynamic single molecule analysis leads to direct visualization of the mechanism of HELQ in DNA double-stranded breaks (DSBs) repair. Check out this application note to learn more.
C-Trap
Molecular biology as never seen before
The C-Trap® provides the world’s first dynamic single-molecule microscope to allow simultaneous manipulation and visualization of single-molecule interactions in real time.

These cards are NOT components because they use the finsweet nested collection logic. To pull in the posible multiple people wo worked on it.
This type of 1-many relation is not supported native in Webflow.
Also this section is hidden when emtpy. To keep everything visible here, that is being done outside the webflow designer from within Slater.
The zinc finger of DNA ligase 3α binds to nucleosomes via an arginine anchor
Coexistence of G-Quadruplex and i-Motif Within a DNA Duplex is Tolerated by a PCBP2-Assisted Replisome
Structure and DNA-bridging activity of the essential Rec114–Mei4 trimer interface
SITC 2025
CAR-TCR Summit 2025
CICON 2025