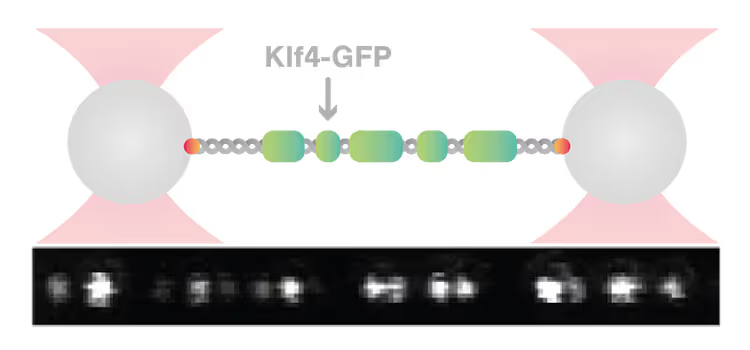
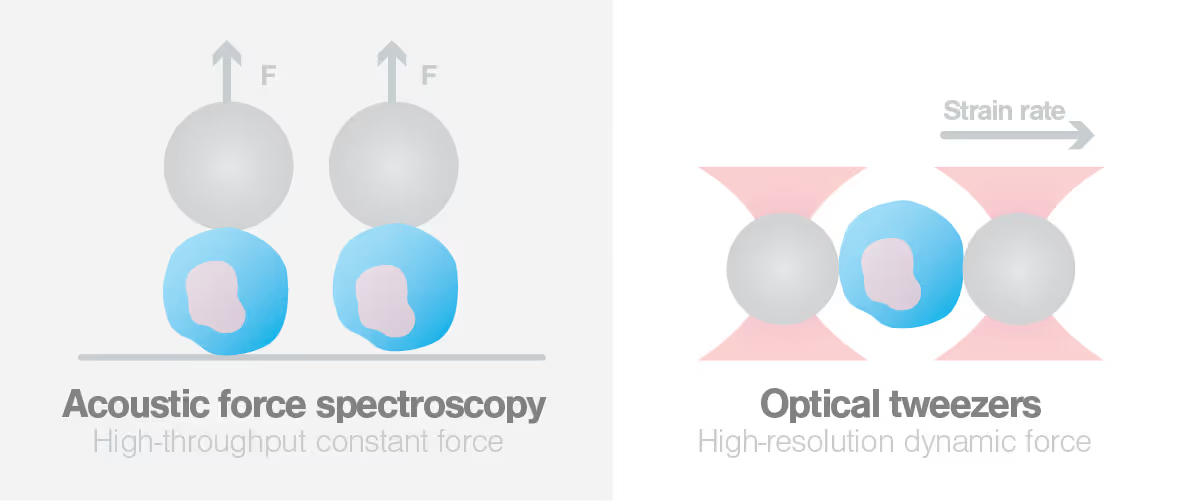
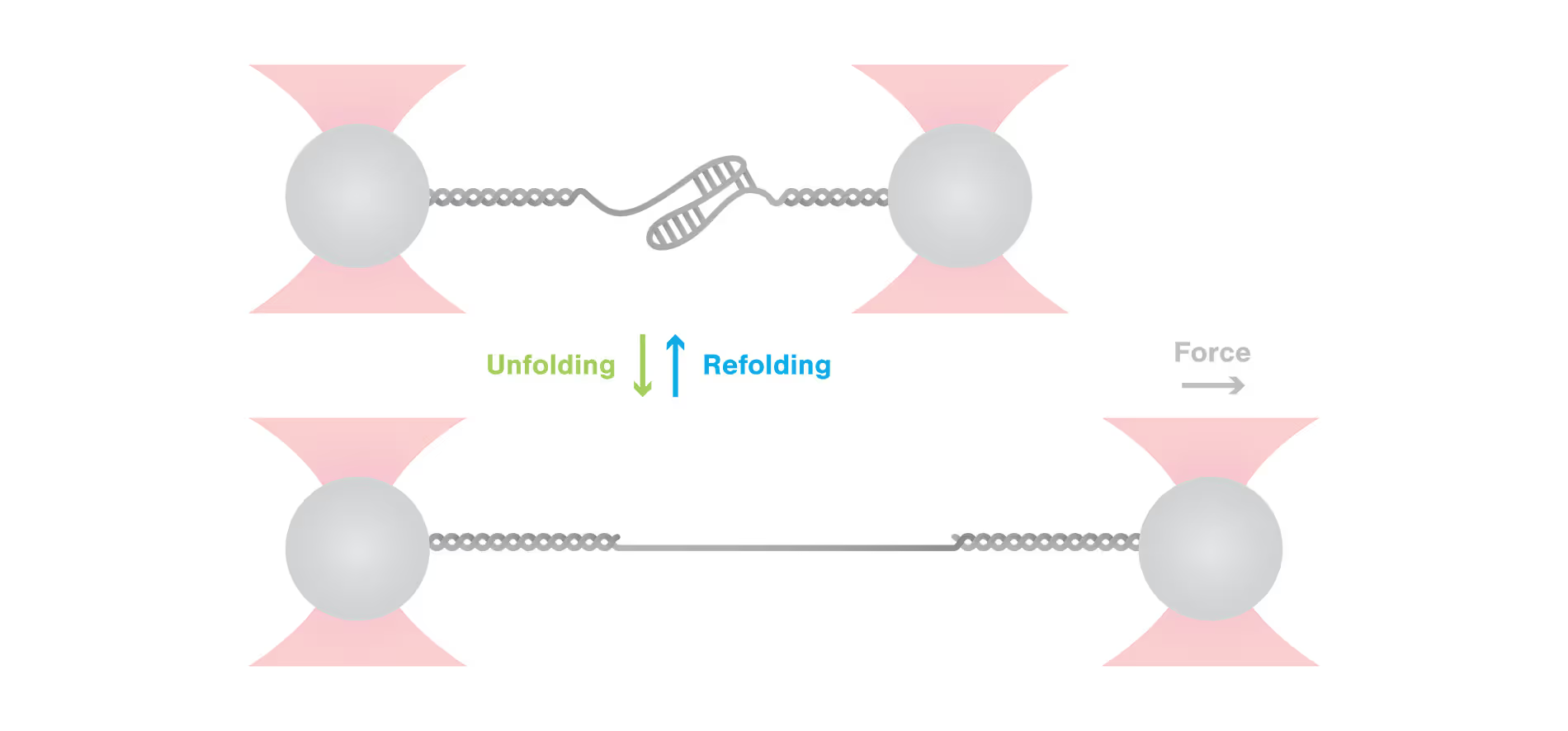
Knowledge Library
Single Molecule Visualisation of Human Topoisomerase 2A Decatenation Reveals Substrate Requirements
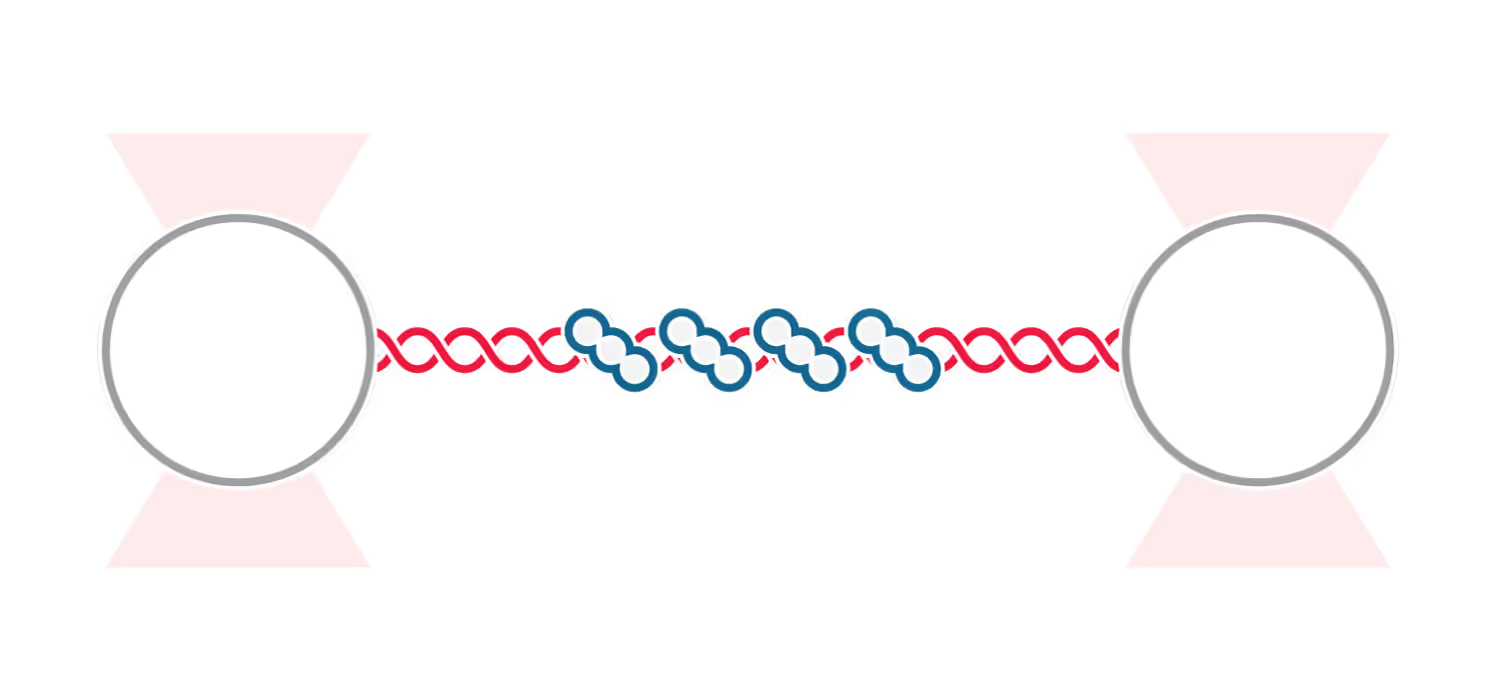
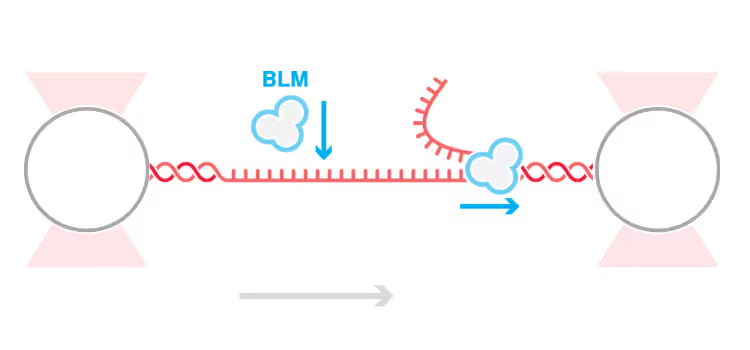
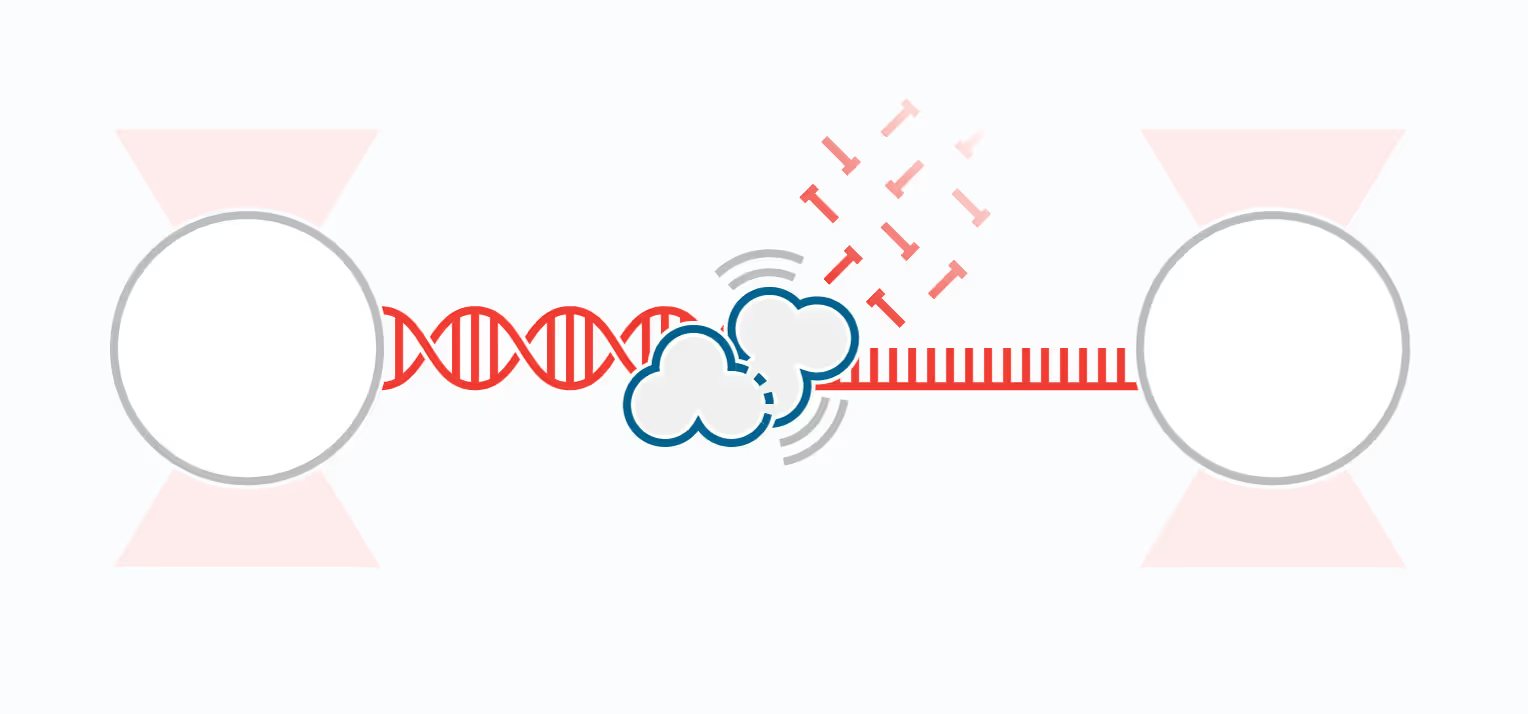
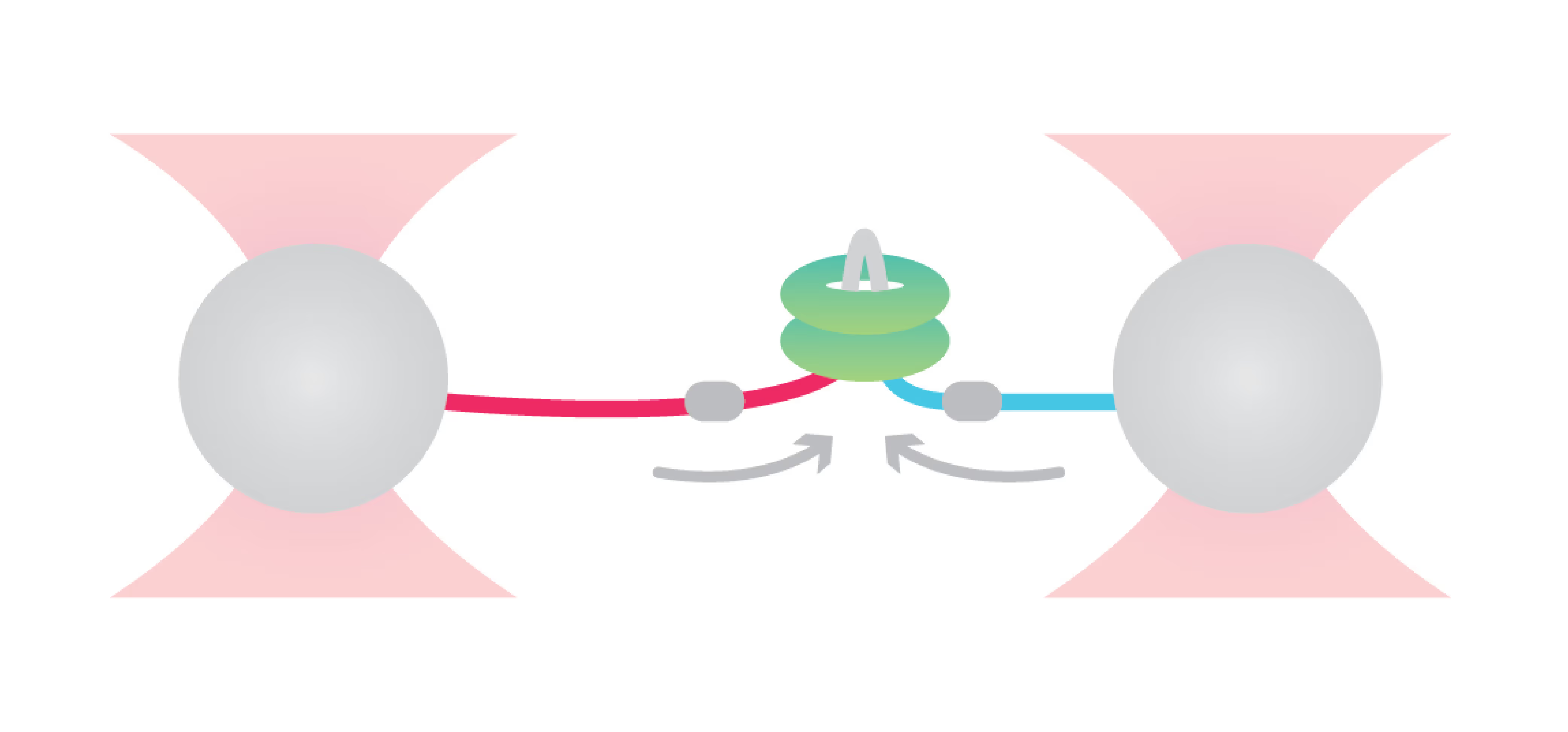
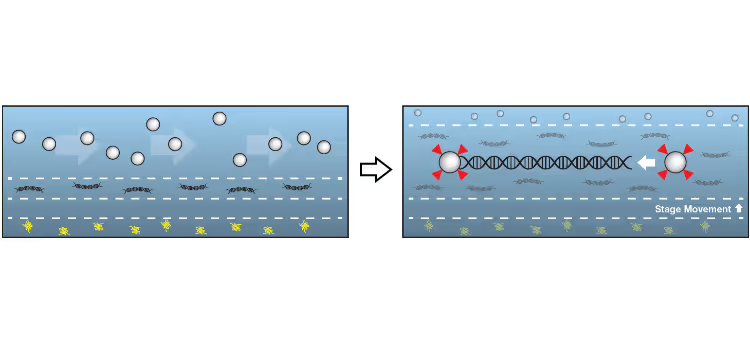
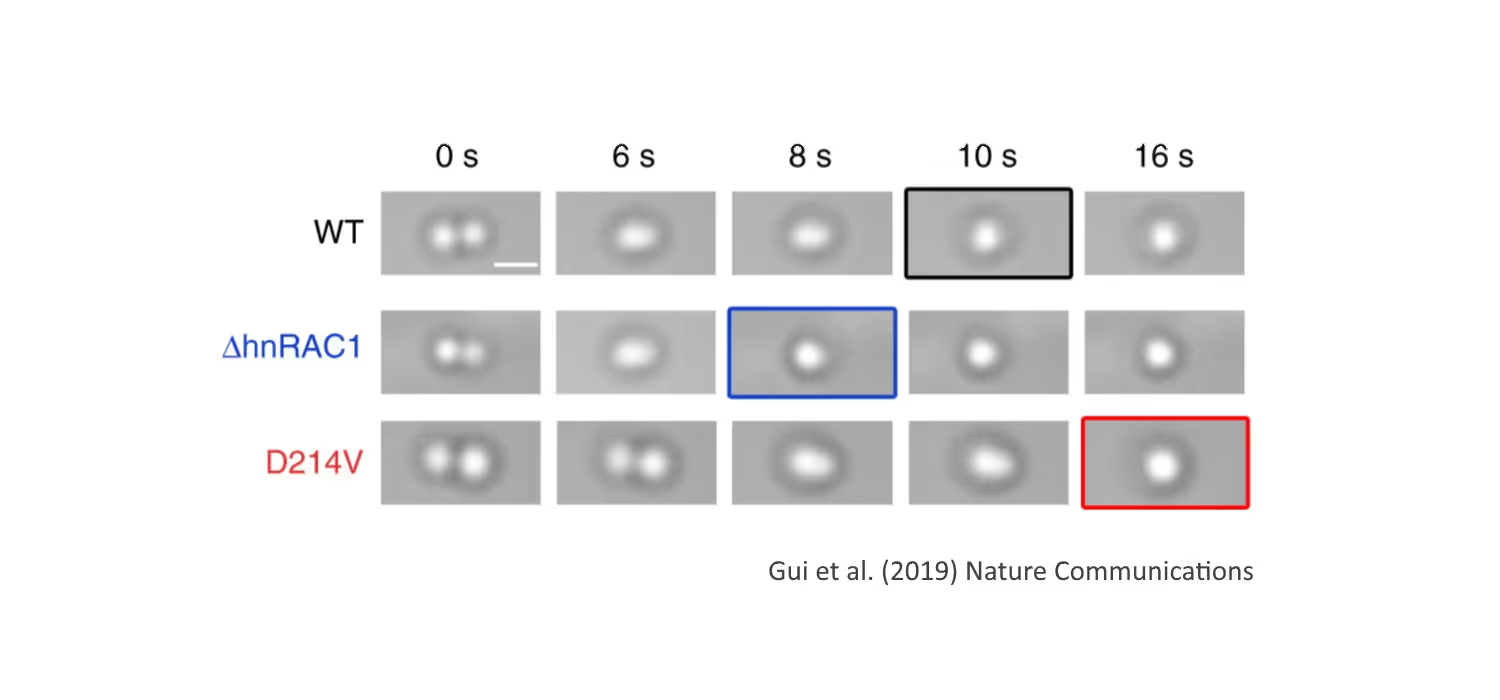
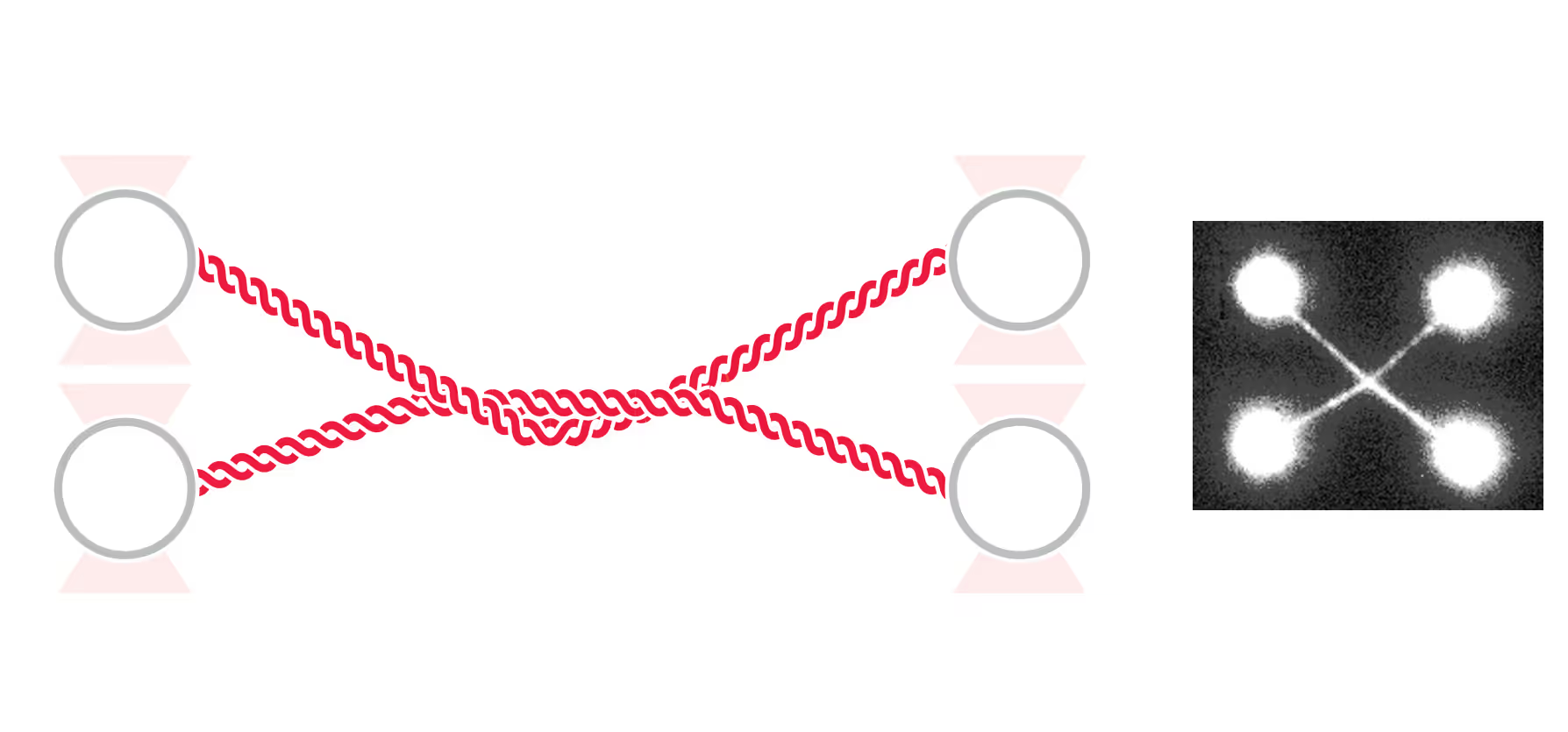
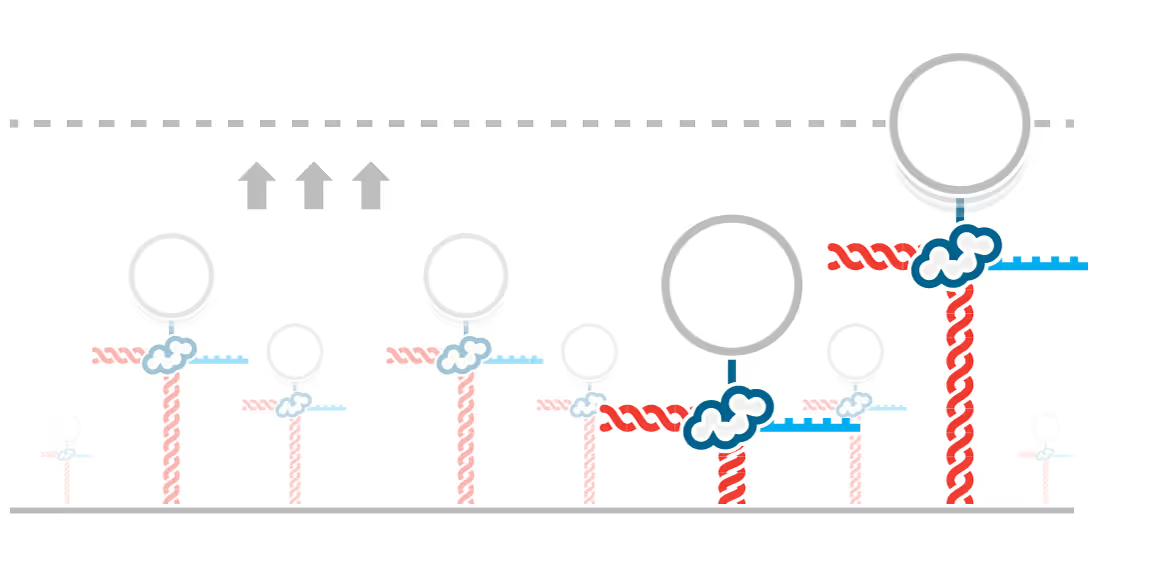
DNA replication introduces double-stranded DNA entanglements, which pose a challenge to cell division and faithful segregation of the genome. During mitosis, Topoisomerase 2A (TOP2A), binds and resolves DNA entanglements by producing a double strand break and then passing the other strand through the break. TOP2A is an essential protein and an important drug target, hence has been extensively studied, but until now the resolution process has not been directly visualised. In this work, I develop an assay for visualising TOP2A activity, mimicking forces that could be applied in a mitotic context, by employing optical tweezers to manually entangle two pieces of DNA(1). I demonstrate that TOP2A DNA resolution is inhibited at high forces, with sharp transition at the half-force of 28 pN. My experiments indicate TOP2A readily binds DNA, however I find resolution is most efficient when TOP2A associates directly at the site of a DNA entanglement. During early mitosis the action of TOP2A chromosome resolution is countered by cohesin and I demonstrate that cohesin readily associated with entangled DNA, and inhibits TOP2A resolution, implicating TOP2A in regulating chromatid cohesion. Collectively, this approach provides novel insights into the important therapeutic target, TOP2A(2).
(1) Meijering, A.E.C., Bakx, J.A.M., Man, T., Heller, I., Wuite, G.J.L., and Peterman, E.J.G. (2022). Implementation of 3D Multi-Color Fluorescence Microscopy in a Quadruple Trap Optical Tweezers System. In Methods in Molecular Biology, pp. 75–100. 10.1007/978-1-0716-2229-2_5.
(2) Cutts, E.E., Saravanan, S., Girvan, P., Ambrose, B., Fisher, G.L.M., Rueda, D.S. and Aragon, L. (2025). Substrate accessibility regulation of human TopIIa decatenation by cohesin. Nature Communications, https://doi.org/10.1038/s41467-025-62505-3
Mechanisms of chromatin remodeler target search and nucleosome mobilization
Chromatin remodelers play a pivotal role in gene regulation by shaping the physical structure of DNA around promoters and regulatory elements. While their biochemical activities are well studied, existing analytical methods fall short in capturing their dynamic activity and the mechanisms by which they search for and engage their targets. This limitation hinders deeper functional assessment, which is essential to the drug development process.
This webcast will explore how individual remodeler proteins including SWR1, RSC, and ISW2 navigate the chromatin landscape by scanning, sliding or hopping to locate and reposition nucleosomes, ultimately influencing access to DNA by transcription machinery. It will feature recent discoveries using single- and dual-color single-molecule tracking with optical tweezers to investigate the one-dimensional (1D) diffusion behaviors of three conserved yeast remodelers—SWR1, RSC, and ISW2—on DNA and sparse nucleosome arrays.
The findings reveal a unified framework for how chromatin remodelers navigate chromatin landscapes to regulate promoter accessibility through coordinated search and remodeling dynamics. They offer new insights into the physical mechanisms underlying gene expression and genome organization, while opening avenues for further exploration of chromatin dynamics and therapeutic strategies.
Kinetic control of mammalian transcription elongation
Transcription elongation by RNA polymerase II (Pol II) is an integral step in eukaryotic gene expression. The speed of Pol II is controlled by a multitude of elongation factors, but the regulatory mechanisms remain incompletely understood, especially for higher eukaryotes. In this work, we developed a single-molecule platform to visualize the dynamics of in vitro reconstituted mammalian transcription elongation complexes (ECs). This platform enabled us to follow the elongation and pausing behavior of EC in real time and dissect the role of each elongation factor in the kinetic control of Pol II. We found that the mammalian EC harbors multiple gears depending on its associated factors and phosphorylation status. The elongation factors are not functionally redundant but act hierarchically and synergistically to achieve optimal EC activity. Such exquisite kinetic regulation may reflect the speed-changing events during the transcription cycle, such as pause-release and termination, and enable cells to adapt to a changing environment.
RNA聚合酶II(Pol II)介导的转录延伸是真核生物基因表达中的关键步骤。虽然已有大量研究表明,Pol II的延伸速度受多种延伸因子的调控,但其具体的调控机制,尤其是在高等真核生物中的调控机制仍未完全被揭示。
本次线上直播讲座,我们有幸邀请到来自美国洛克菲勒大学刘诗欣组的王昱焜博士,分享其在单分子水平重建哺乳动物转录延伸复合物(EC)平台方面的突破性研究成果。该研究利用LUMICKS C-Trap实时观察EC的延伸与停顿过程,揭示各类延伸因子如何协同调控Pol II动力学状态,推动转录过程的精细调节。
C-Trap Accelerator Suite
Get an inside look at the new C-Trap Accelerater Suite and see how it can transform your experiments. Our product managers will walk you through the powerful new features, which are designed to make your C-Trap faster, easier to use, and more reproducible than ever.
A self-enforcing protein-DNA surface hydrogel safeguard nuclear integrity
The nuclear envelope protects the genome from mechanical stress during processes such as migration, division, and compression , but how it buffers forces at the scale of DNA remains unclear. Here, we utilize optical tweezers to show that a multivalent protein–DNA co-condensate containing the nuclear envelope protein LEM2 and the DNA-binding protein BAF shield DNA beyond its melting point at 65 pN. Under load, their collective assembly induces an unconventional DNA stiffening effect that provides mechanical reinforcement, dependent on the intrinsically disordered region (IDR) of LEM2. Within cells, these components form an elastic surface hydrogel at the nuclear periphery, visible as a continuous surface by cryo-electron tomography. Disruption of this surface hydrogel increases DNA damage and micronuclei formation during nuclear deformation. Together, this work expands the functional repertoire of condensates, revealing a load-responsive nuclear surface hydrogel at the mesoscale that mitigates mechanical stress.
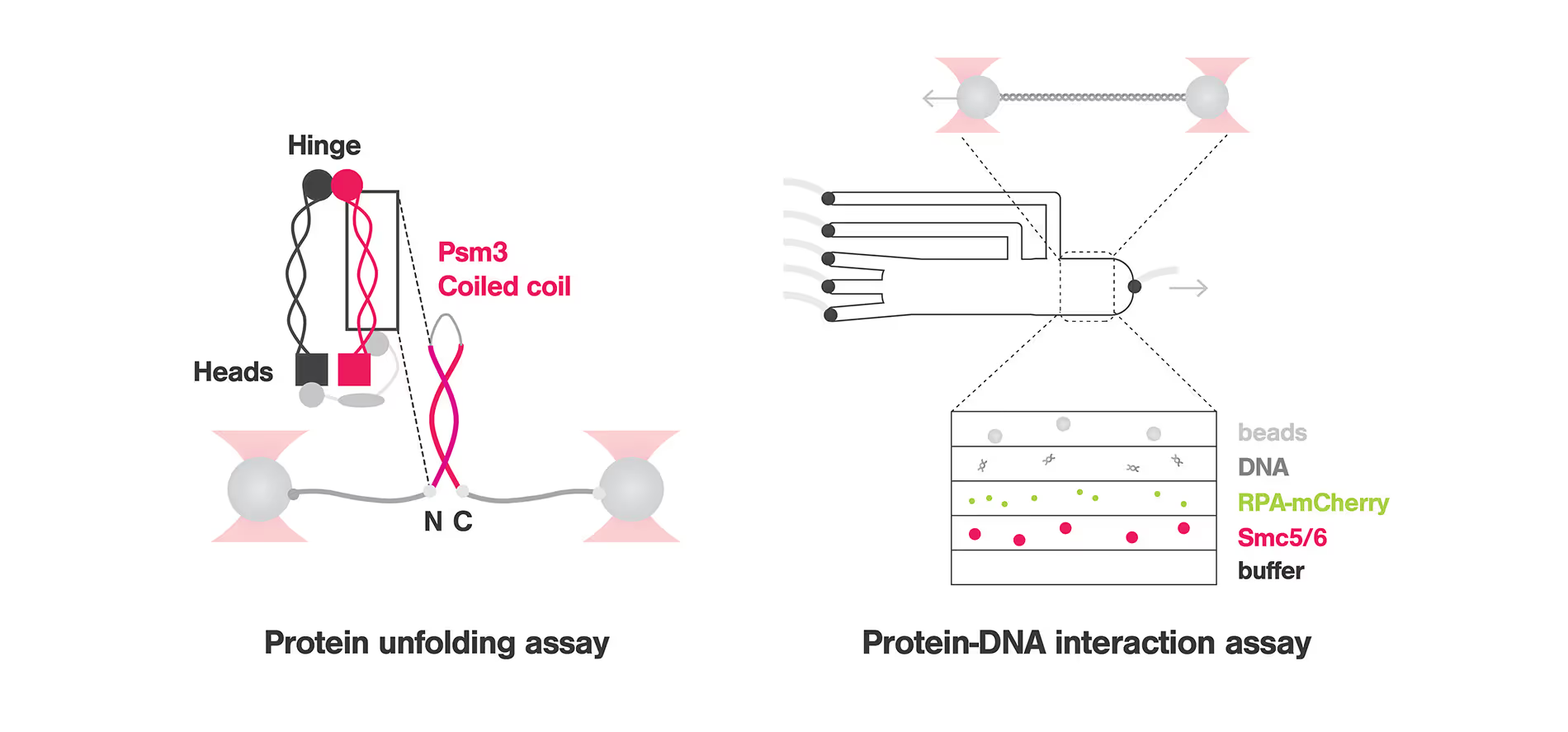
New Insights in Chromosome Organization: Single Molecule Analysis of Eukaryotic SMC Complexes
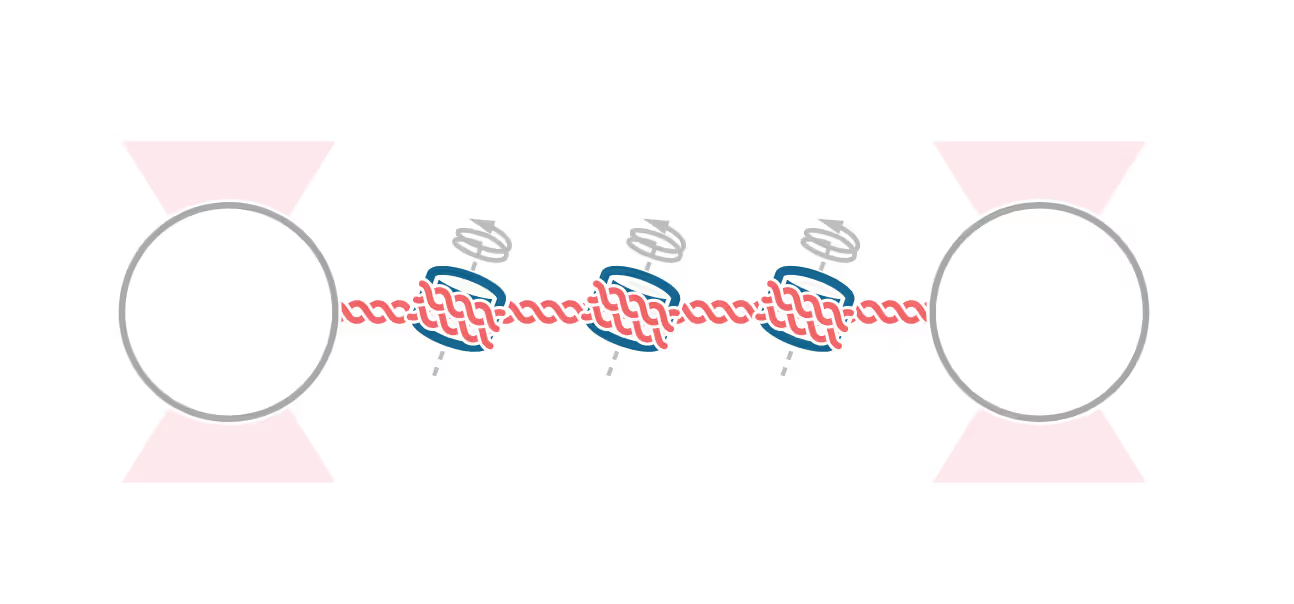
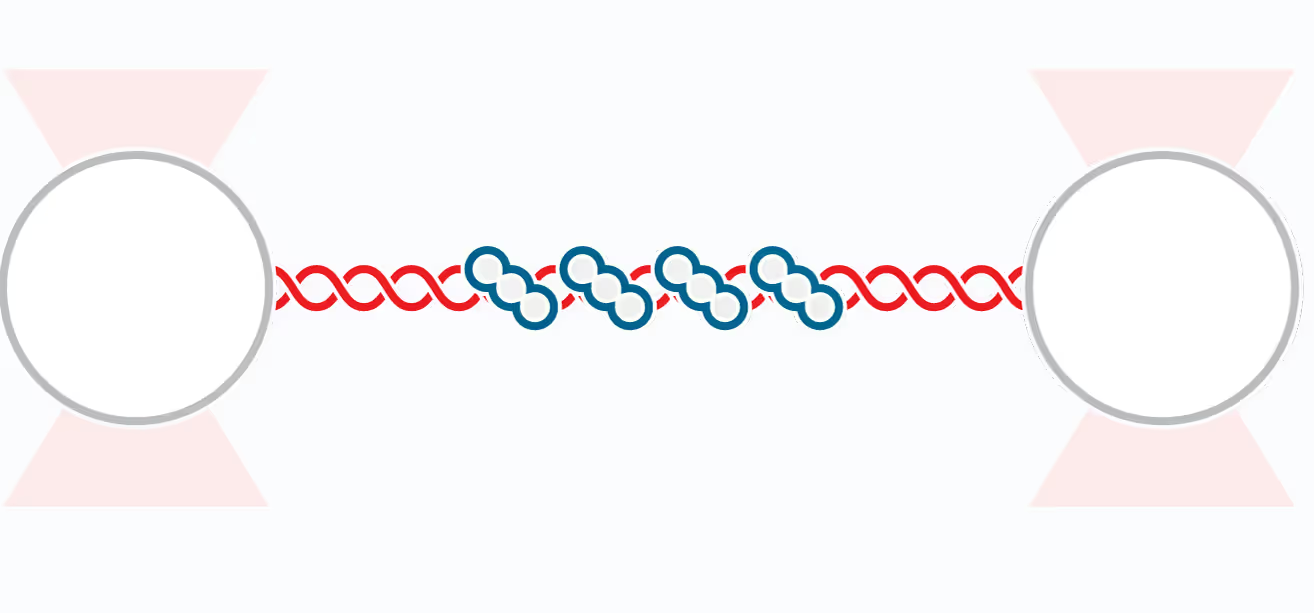
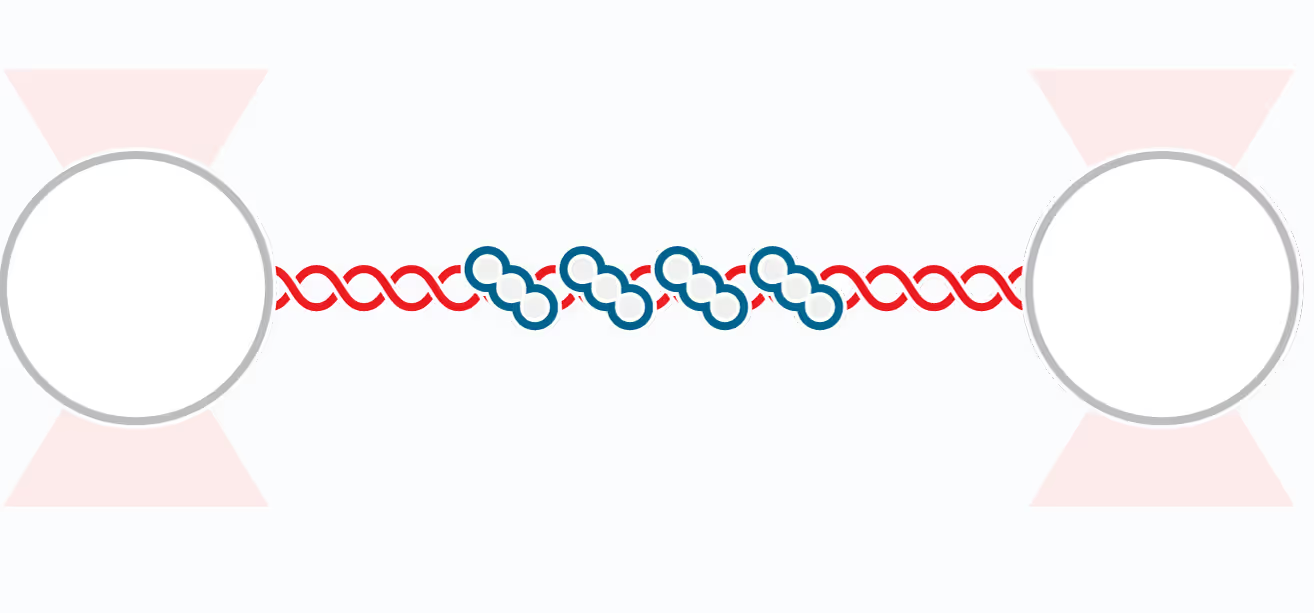
Structural Maintenance of Chromosome (SMC) complexes, cohesin, condensin and Smc5/6, play a fundamental role in genome organization, facilitating chromosome compaction, segregation, and DNA repair. Despite their essential functions, the mechanisms by which the complexes interact with different DNA substrates and influence topological transitions remain not fully understood. Using the LUMICKS C-Trap, we have employed single-molecule approaches to analyze the behavior of purified SMC complexes, focusing on yeast cohesin and human SMC5/6, on different DNA substrates, including double-stranded (dsDNA) and single-stranded DNA (ssDNA). Additionally, we have used a quadrupole optical trap to bridging by SMC complexes and their effect on DNA decatenation by Topoisomerase IIα (Top2A). These findings provide new insights into the fundamental properties and requirements of cohesin and Smc5/6’s interaction with DNA substrates, as well as their ability to bridge two independent DNAs.
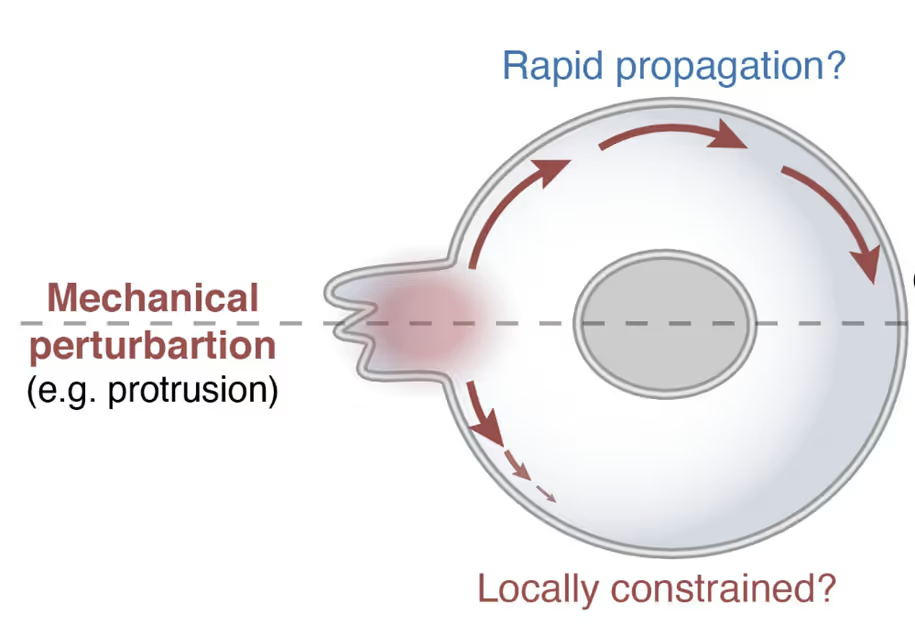
Live cell force dynamics – do cell membranes support or resist tension propagation?
What are the rules of membrane tension propagation in cells? For proper function, cells need to link short-range biochemical signaling events with long-range integration of cell physiology. Forces transmitted through the plasma membrane are thought to serve as this globally integrator. However, conflicting observations have left the field divided as to whether cell membranes support or resist tension propagation. This discrepancy likely originates from the use of exogenous forces that may not accurately mimic endogenous forces. We overcome this complication by leveraging optogenetics to directly control localized actin-based protrusions or actomyosin contractions while simultaneously monitoring the propagation of membrane tension using dual-trap optical tweezers. Surprisingly, actin-driven protrusions and actomyosin contractions both elicit rapid global membrane tension propagation, whereas forces applied to cell membranes alone do not. We present a simple unifying mechanical model in which mechanical forces that engage the actin cortex drive rapid, robust membrane tension propagation through long-range membrane flows.
Breakthroughs into molecular machines on DNA and chromatin
Genome replication and gene expression are carried out by macromolecular machines that exist at nanometer scale and generate piconewton forces. Challenged by the hierarchical chromatin organization and omnipresent thermal fluctuations, these DNA-based machines still accomplish their tasks with remarkable efficiency and accuracy. We leverage single-molecule techniques, particularly correlative fluorescence and force microscopy (smCFFM), to probe the dynamics and mechanics of replication, transcription, and chromatin machinery. These investigations have yielded new insights into the principles of genetic and epigenetic inheritance.
FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions
DNA crosslinks block DNA replication and are repaired by the Fanconi Anemia pathway. The FANCD2-FANCI (D2-I) protein complex is central to this process as it initiates repair and is also known to play a more general role in DNA repair and in protecting stalled replication forks from unscheduled degradation.
Here, using single-molecule imaging, we investigate the behaivior of D2-I and provide a unified molecular mechanism that reconciles the roles of D2-I in recognition and protection of stalled replication forks in multiple DNA repair pathways.
Decoding the onset of Amyotrophic Lateral Sclerosis (ALS): Liquid-liquid phase transition of Intrinsically Disordered Proteins
Join us for an insightful webinar where we delve into the fascinating world of Intrinsically Disordered Proteins (IDPs), their complex phase behavior, and the implications for neurodegenerative disorders like Amyotrophic Lateral Sclerosis (ALS). Brought to you by Priya Banerjee Lab from the University at Buffalo, we will explore the enigmatic properties of IDPs, which account for a significant portion of the eukaryotic proteome, and challenge the classical protein structure-function paradigm.
The webinar will feature a particular focus on Fused in Sarcoma (FUS) protein, a type of RNA-binding protein that forms biomolecular condensates or ‘droplets’ via phase separation. FUS droplets are known to play significant roles in cellular functions such as DNA repair, RNA metabolism, and transcription regulation. In the context of ALS, mutations in the FUS gene can result in abnormal protein behavior, including the formation of aberrant, persistent FUS droplets that are associated with neurodegeneration.
In addition to illuminating the role of FUS droplets in ALS, we will dissect the phase behavior of Protein-RNA condensates, including how RNA binding regulates their phase behavior, compositional specificity, and transport properties. By employing advanced techniques like fluorescence microscopy and optical tweezers, we’ll gain a quantitative understanding of the molecular driving forces that underlie these critical processes.
Furthermore, the session will delve into the biophysics of phase transitions, genome packaging, and the organization of the genome into membrane-less compartments to regulate gene expression. This in-depth exploration promises unique insights into the complex world of IDPs, biomolecular condensates, and their implication in ALS. Join us for this enlightening journey into one of biology’s most intriguing fields.
A deep dive into Nucleotide Excision Repair (NER) and its crucial role in Alkyl-DNA lesion repair and cancer prevention
In this session, hosted by DNA repair expert Dr. Ingrid Tessmer, Rudolf Virchow Center for Experimental Biomedicine, University of Würzburg, we dive deep into the role of alkyltransferase-like proteins (ATLs) and their role in NER. Despite their inherent catalytic inactivity, ATLs play a remarkable role in targeting alkyl lesions for repair by the NER system. Through a combination of single-molecule and ensemble methodologies, a detailed view of the recruitment process of UvrA – the initiating enzyme of prokaryotic NER – to an alkyl lesion by ATL has been observed for the first time.
Moreover, we delve into the mechanisms of lesion recognition by ATL, and illustrate the dynamic DNA lesion search undertaken by highly active ATL and ATL-UvrA complexes.
Don’t miss this opportunity to broaden your understanding of DNA repair and its potential role in revolutionizing cancer treatment strategies.
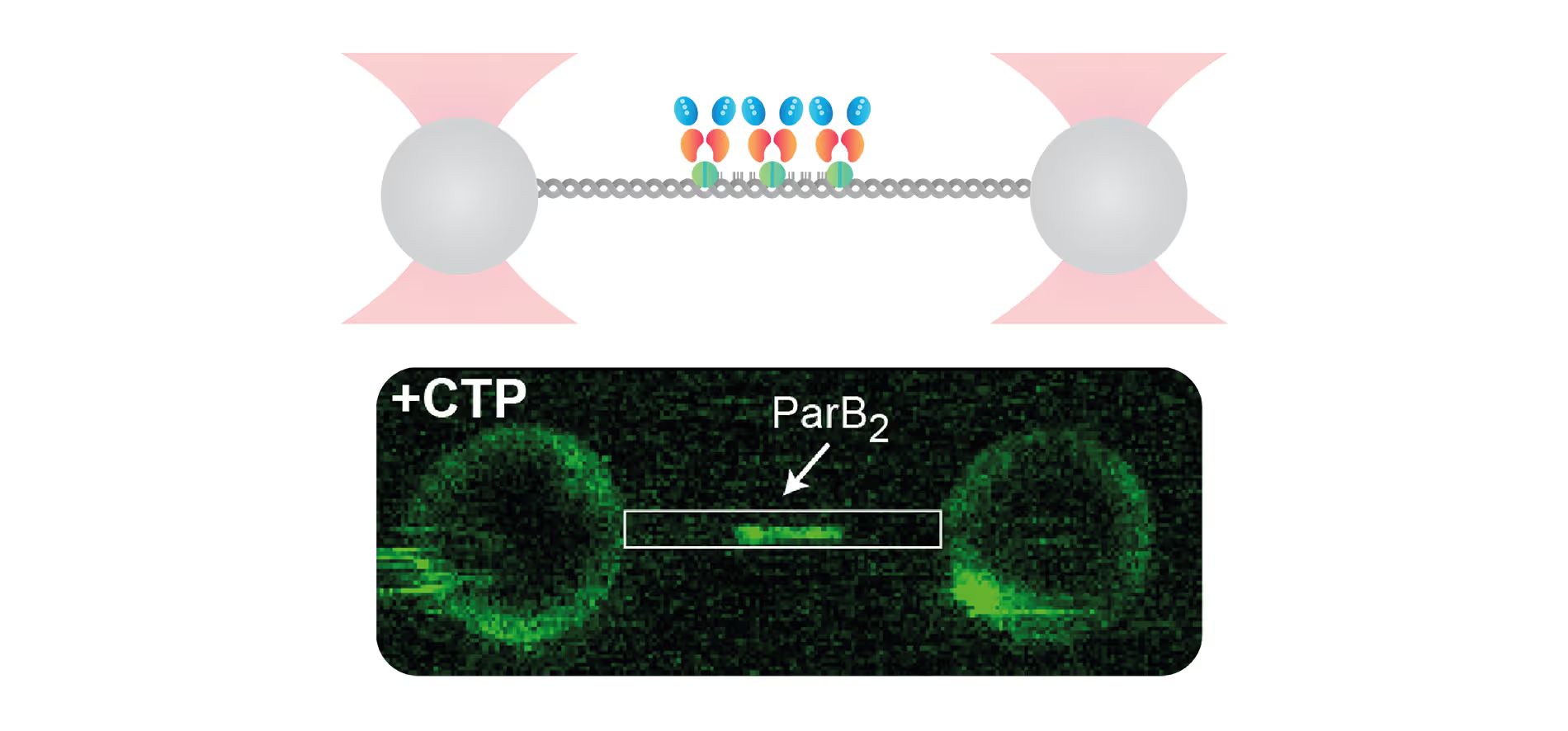
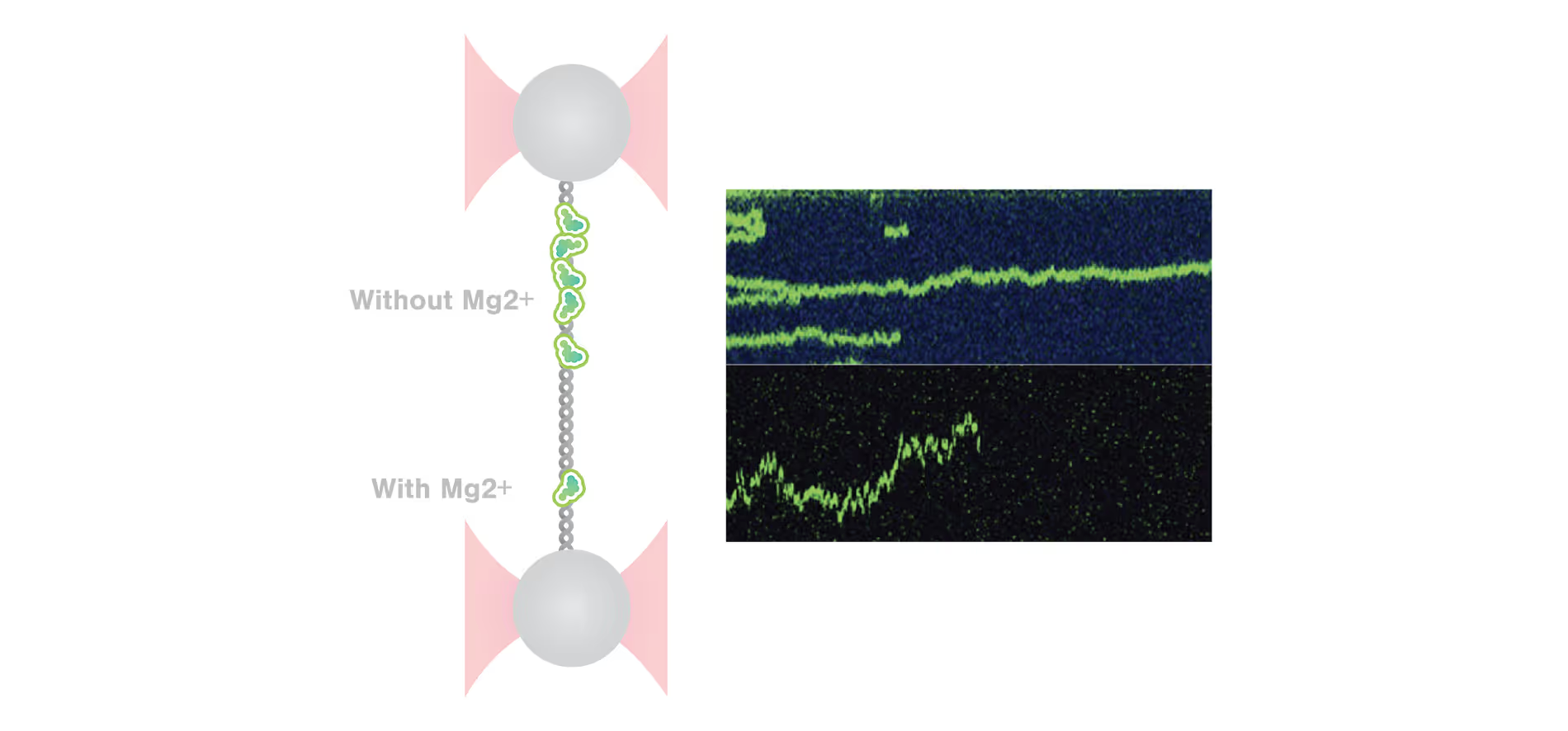
Insights into Bacterial DNA Condensation and chromosome organization
Guided by our host, a renowned expert in bacterial cell biology, Dr. Fernando Moreno Herrero from the Department of Macromolecular Structures, Centro Nacional de Biotecnología, Consejo Superior de Investigaciones Científicas, Madrid, we will explore the critical ParABS partitioning system and the SMC complex, which serve as the driving forces behind bacterial chromosome segregation.
Using cutting-edge single-molecule techniques and optical tweezers combined with a confocal microscope, we offer an exclusive view into how cytidine triphosphate (CTP) binding and hydrolysis play integral roles in the interaction between parS and ParB.
Unlocking the power of Cas12a: Novel insights into engineered endonucleases for safe and precise next generation genome editing
The advent of powerful and precise gene-editing tools is transforming the way we approach health, medicine, and biological research, opening up possibilities that were once considered science fiction.
Join us today in this webinar to unravel the secret of Cas12a endonuclease, a component of the revolutionary CRISPR-Cas gene-editing technology. Prof. Guillermo Montoya will guide us through his groundbreaking research that explores how Cas12a, an RNA-guided enzyme, interacts with bacteriophage λ-DNA. This journey into the world of genome editing will not only deepen our understanding of this complex tool but also inspire us to imagine the potential advancements in biomedicine and biotechnology.
As we embark on this journey together, we are excited to share and explore the novel insights and possibilities that are becoming accessible through these advancements in gene-editing technologies. Let’s dive in and explore the extraordinary world of Cas12a and its implications in gene editing.
Single-molecule analysis of cancer DNA-protein interactions from nuclear extracts
Understanding of DNA repair mechanisms could advance treatments for cancer and diseases of aging. But reconstituting DNA repair protein complexes from cancerous tissues to study their mechanisms of action is often time-consuming or, in some cases, impossible. A new technique performing dynamic single-molecule analysis directly on nuclear extracts allows rapid mechanistic analysis of mutant proteins from cancer cells, providing previously unseen insights into their mechanisms of action. This new innovative tool, when combined with rapid data analysis, represents a bridge between the study of biochemistry of purified proteins and molecular biology.
Studying DNA and chromatin replication using integrated force-fluorescence microscopy
Chromatin replication is a highly complex process and is crucial for genome integrity, and thus the proper functioning and survival of any organism. Copying chromatinized DNA with its sophisticated structural elements like nucleosomes and structural maintenance of chromatin (SMC) protein-induced loops as well as various modifications and possible damage sites poses quite a challenge to the molecular replication system (replisome).
In this webinar, Prof. Nynke Dekker explains how she and her team have used Dynamic Single-Molecule (DSM) microscopy to identify and quantify fundamental molecular interactions between the origin recognition complex (ORC) and minichromosome maintenance protein (MCM) complex in the context of nucleosomes.
Linking Mechanical Stability with in vivo Recombination: Single-molecule Research Reveals Bacterial Antibiotic Resistance
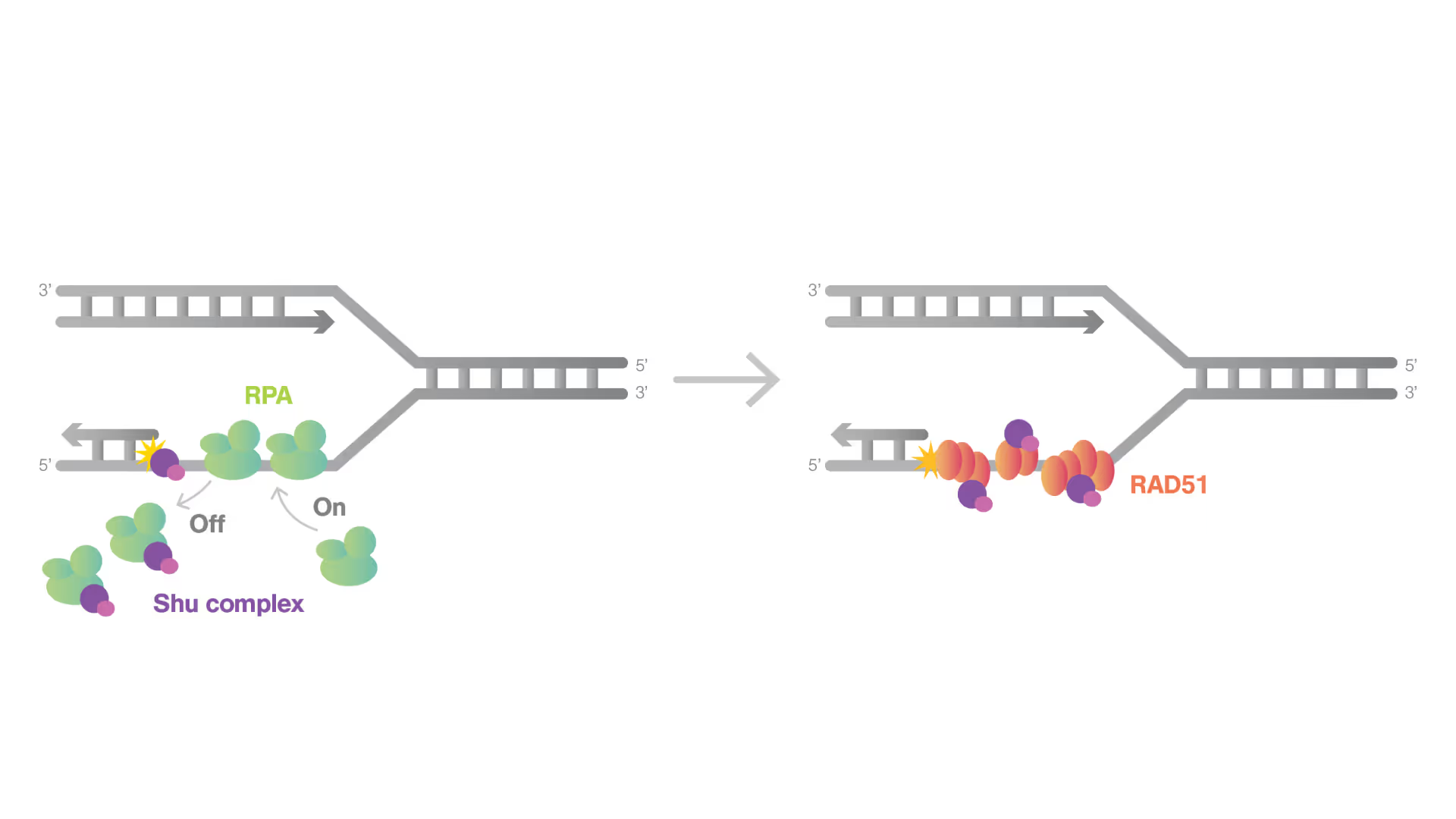
Shu Complex: A New Frontier in DNA Repair and Cancer Therapy
DNA Polymerase Undergoes Rapid Exchange and Retains Memory at Replication Forks
Key mechanism in Fanconi Anemia pathway discovered with the C-Trap
Chromatin remodeling dynamics revealed at the single-molecule level in a recent pioneering study
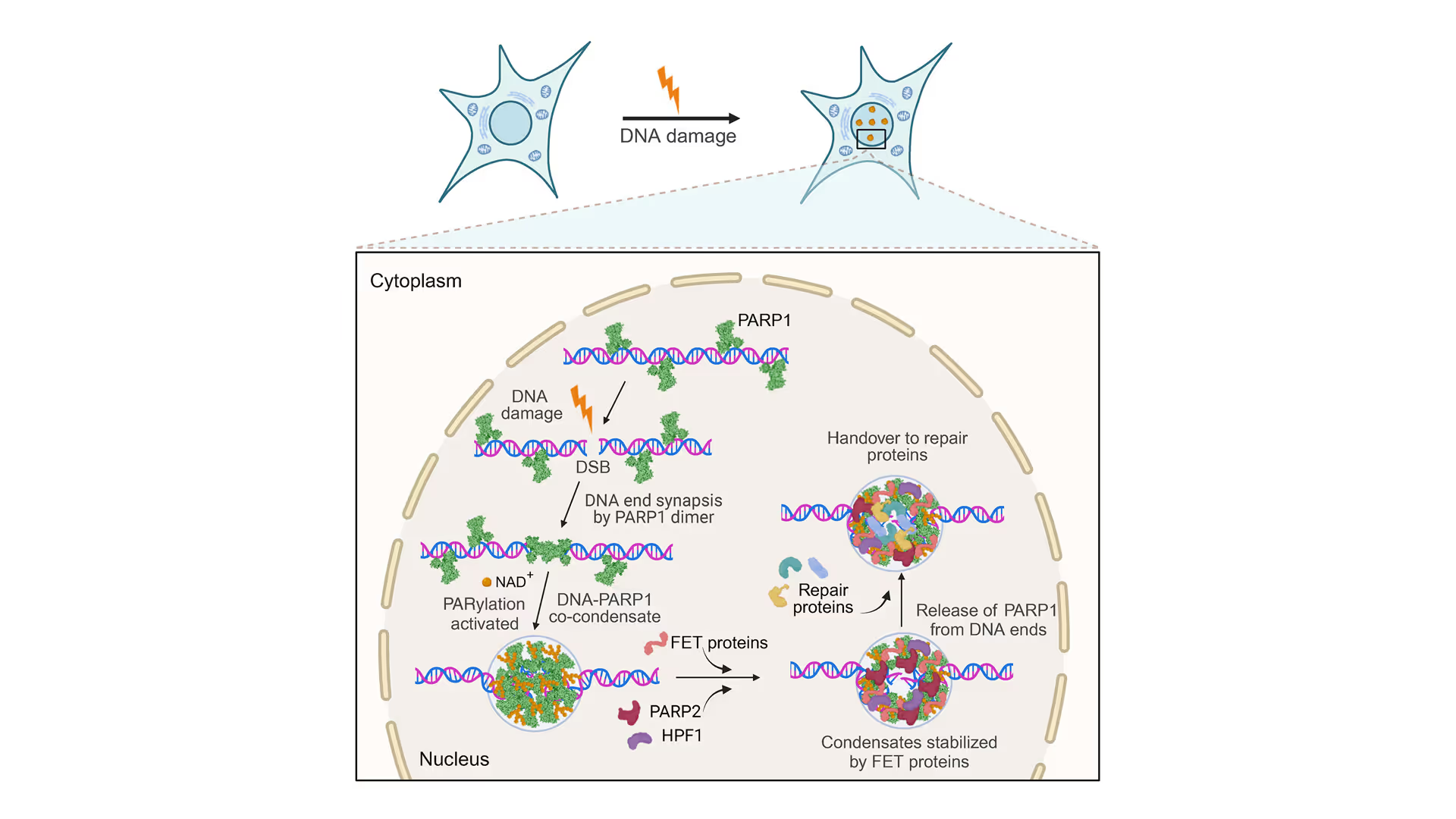
Securing Broken DNA Ends: PARP1’s Vital Role in Co-Condensation with DNA
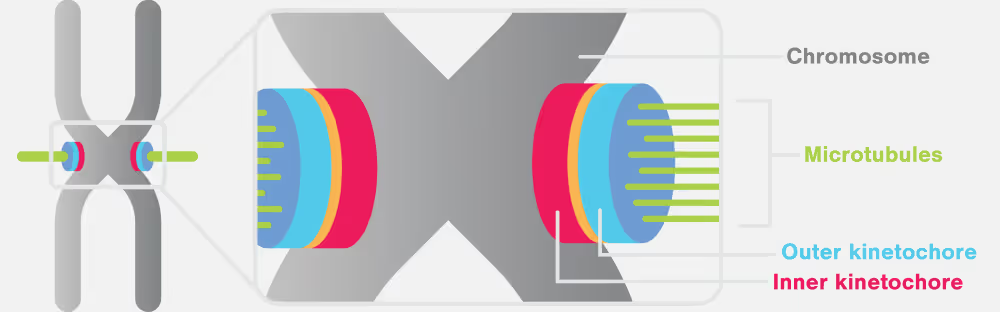
Unveiling Key Mechanisms in Kinetochore-Mediated Chromosome Segregation with the C-Trap
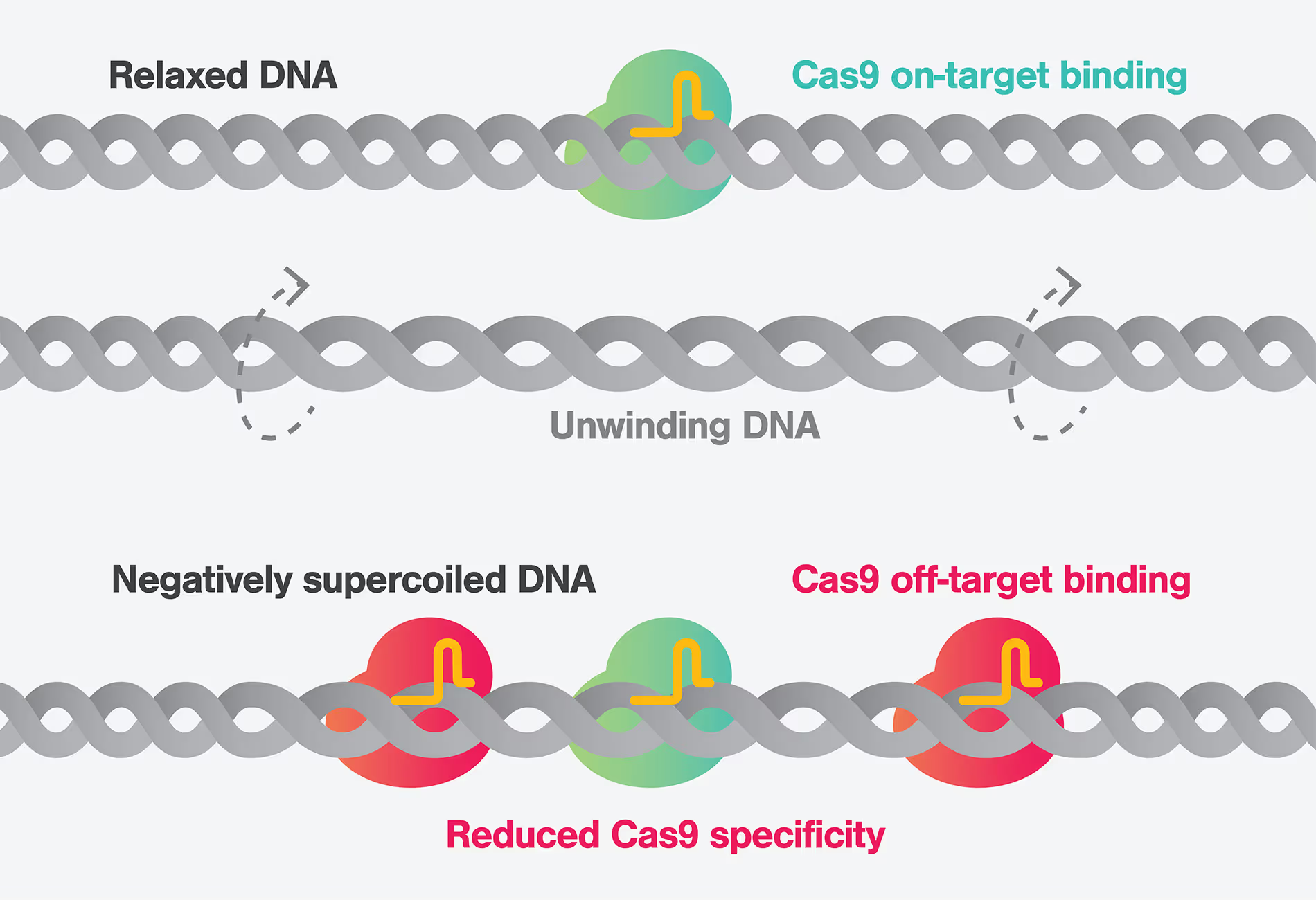
Towards safe and precise gene therapies: revealing the impact of DNA Supercoiling on CRISPR-Cas9 specificity
Cell mechanics with the C-Trap – cellular tension propagation revisited
The C-Trap uncovers new insights into the mechanisms for ATP-dependent chromatin remodeling
C-Trap experiments reveal novel mechanism for transcription termination in bacteria
100th study using the C-Trap® provides crucial insights into how the Smc5/6 complex stabilizes certain DNA structures
C-Trap experiments shed light on the mechanisms that govern Structural Maintenance of Chromosome (SMC) Complexes
Unlocking the secrets of DNA-binding proteins in nuclear extracts using dynamic single-molecule analysis
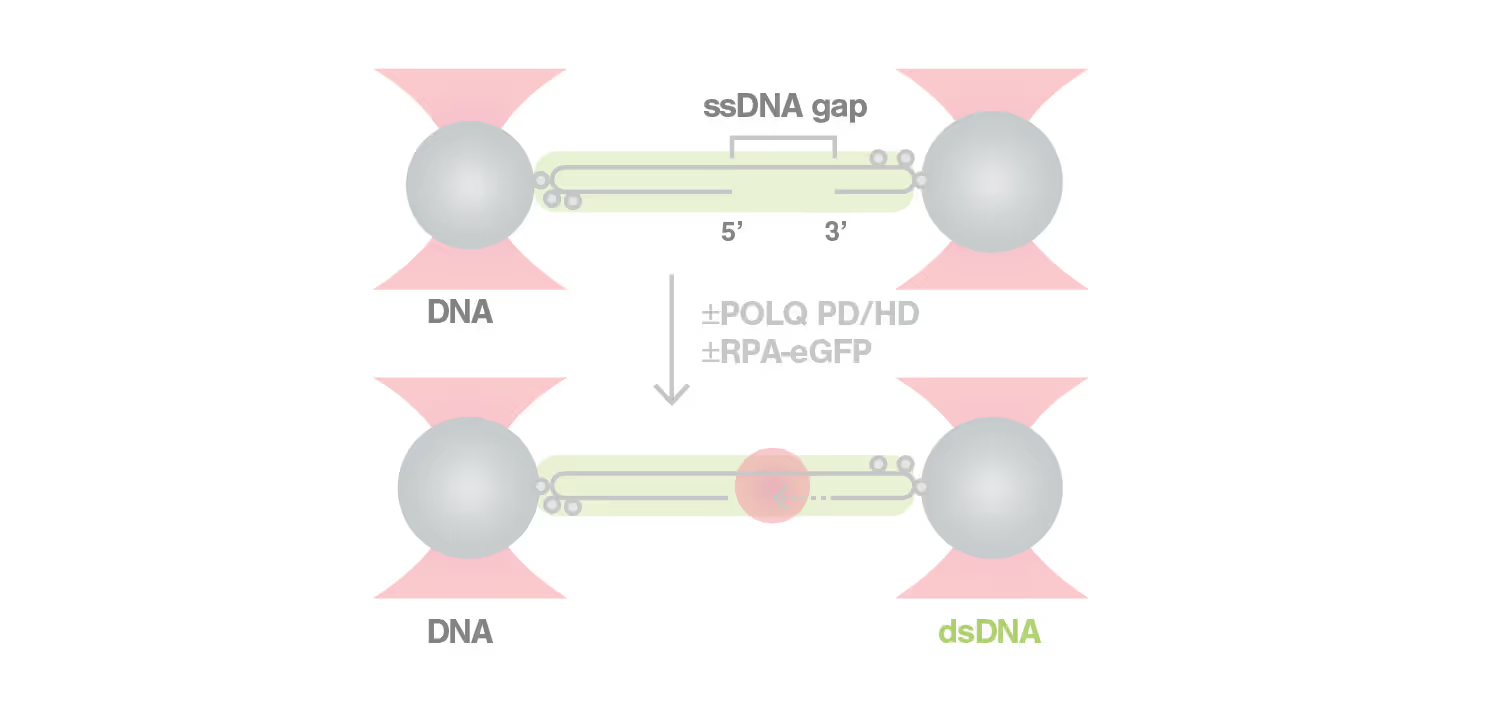
C-Trap®experiments highlight the potential of POLQ inhibitors as BRCA-deficient cancer therapeutics
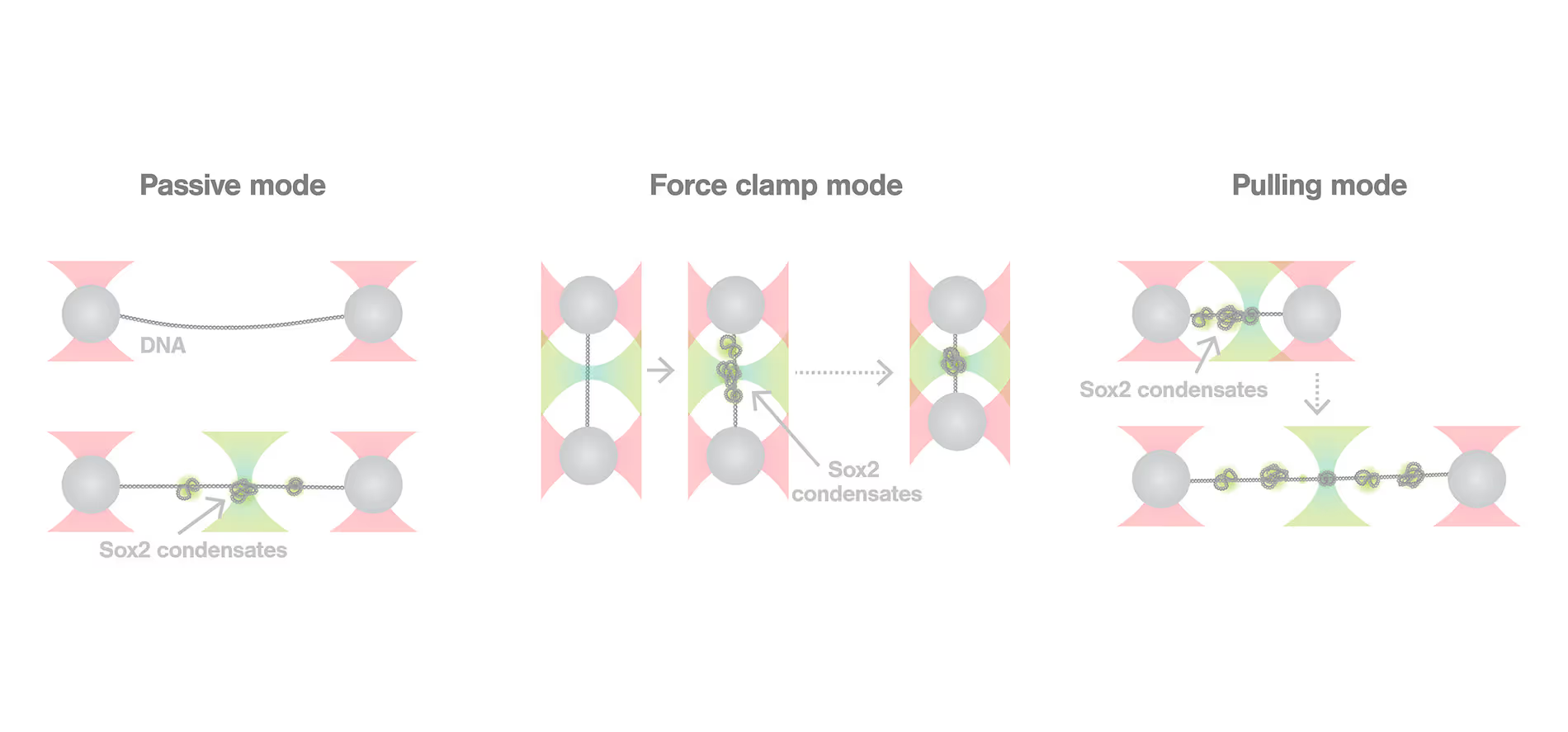
Transcription factors and chromatin co-condensate and generate forces that alter genomic structure and function
Dynamic single-molecule experiments expose catch bonds as key to cell strength and flexibility
C-Trap experiments suggest tubulin lattice state dictates tau envelope formation
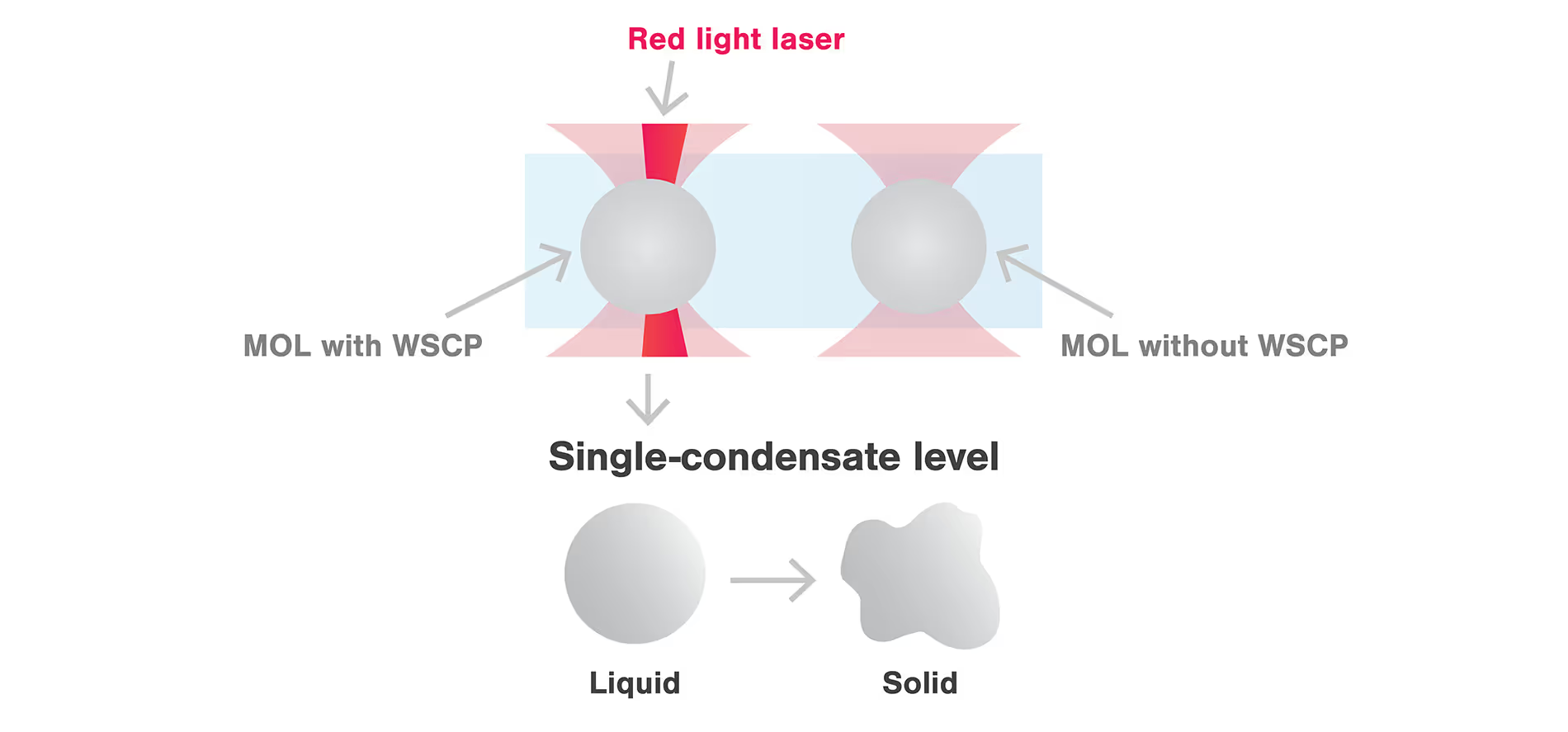
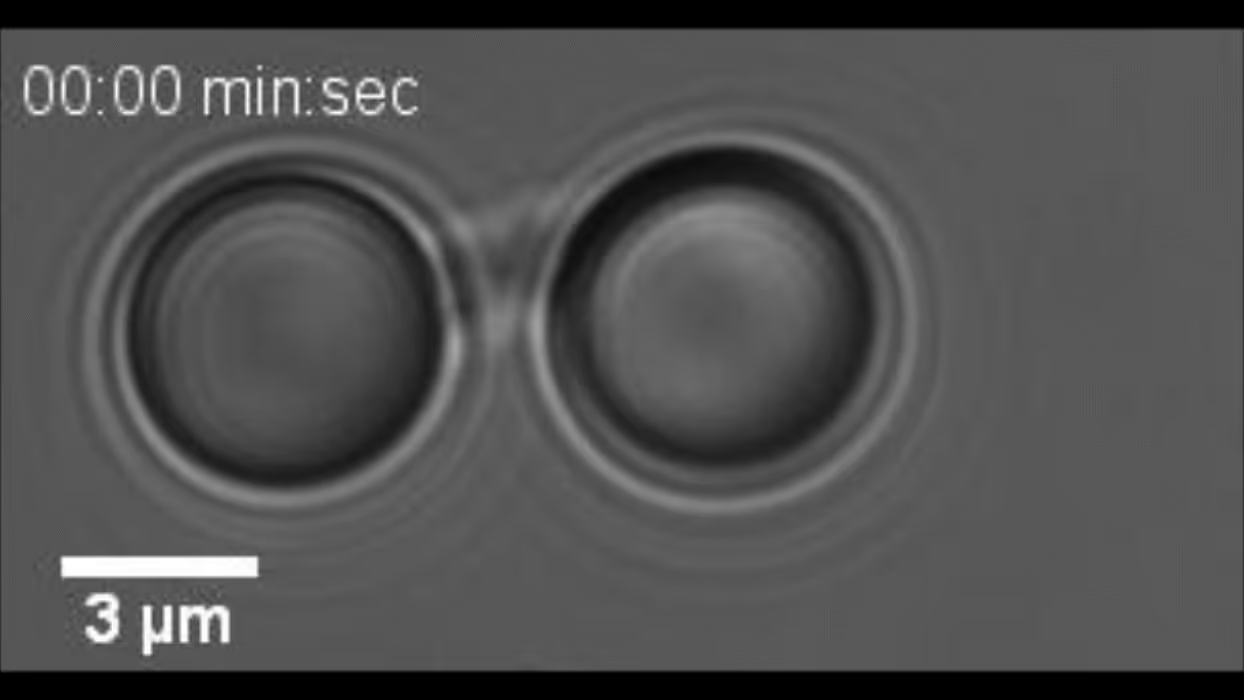
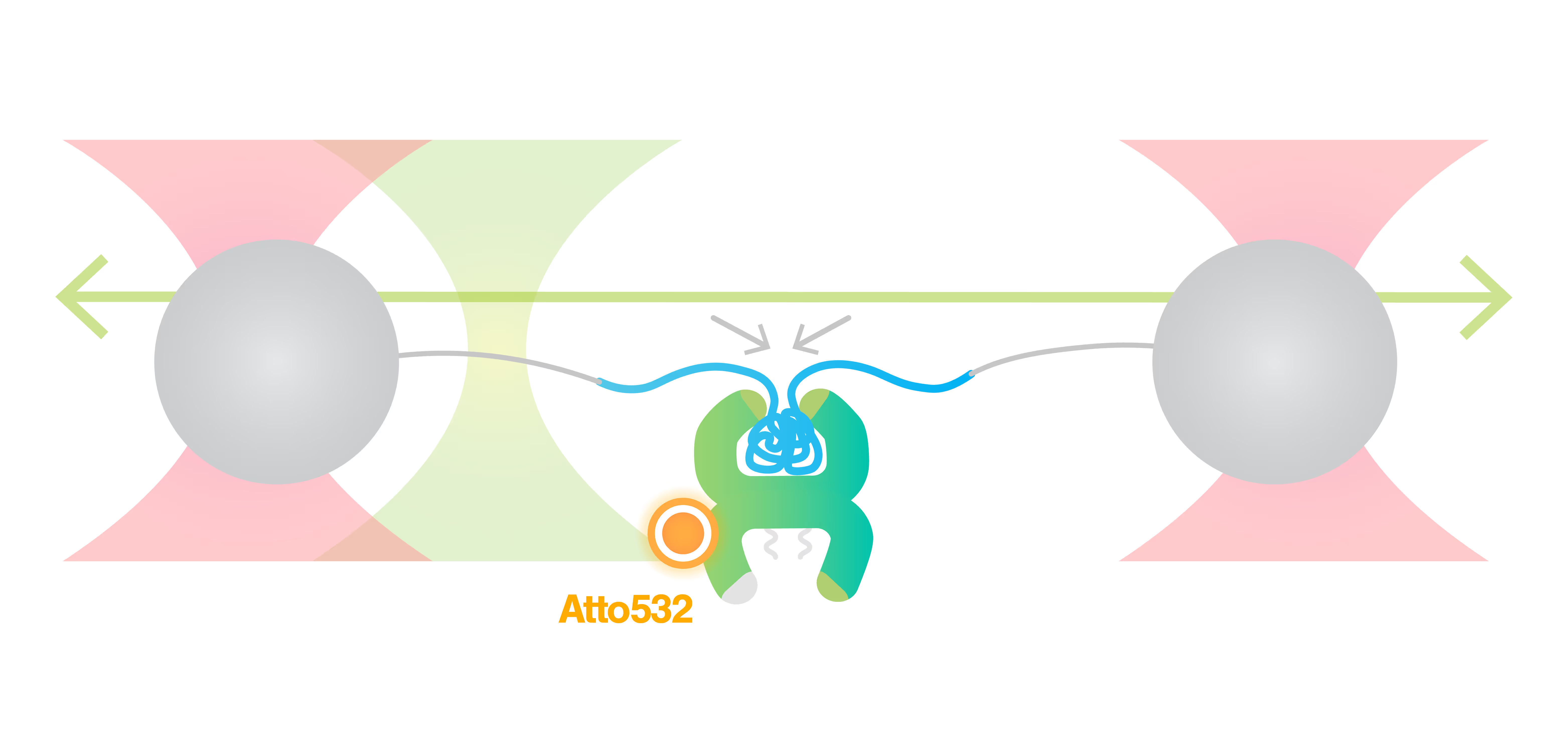
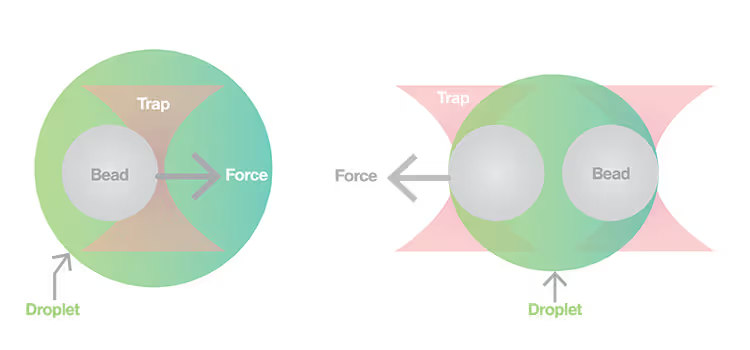
m-Trap allows precise control of biological activities in synthetic membraneless organelles (MLOs) in real time
Dynamic single-molecule analysis reveals non-linear stiffening of mitotic chromosomes
C-Trap studies reveal GroEL-ES protein folding acceleration mechanism
Exploring biomolecular condensate dynamics with the C-Trap®reveals viscoelastic fluid behavior
C-Trap experiments reveal mechanism of protein-DNA co-condensate formation
C-Trap® experiments reveal how a pioneer transcription factor interacts with DNA to regulate gene expression
Probing monocyte forces with the C-Trap®and acoustic force spectroscopy reveals CCL2 regulation of monocyte mechanics
C-Trap experiments contribute to elucidating the role of the helicase HELQ in different mechanisms of DNA repair
ZAP-S binding prevents the refolding of SARS-CoV-2 RNA, inhibits programmed ribosomal frameshifting
ParB sliding to non-specific DNA regions visualized using C-Trap®
Using C-Trap to unravel different binding modes of architectural DNA-binding proteins at different ionic conditions and DNA conformation
Insights into the mechanism of Cas12a dynamics using C-Trap®
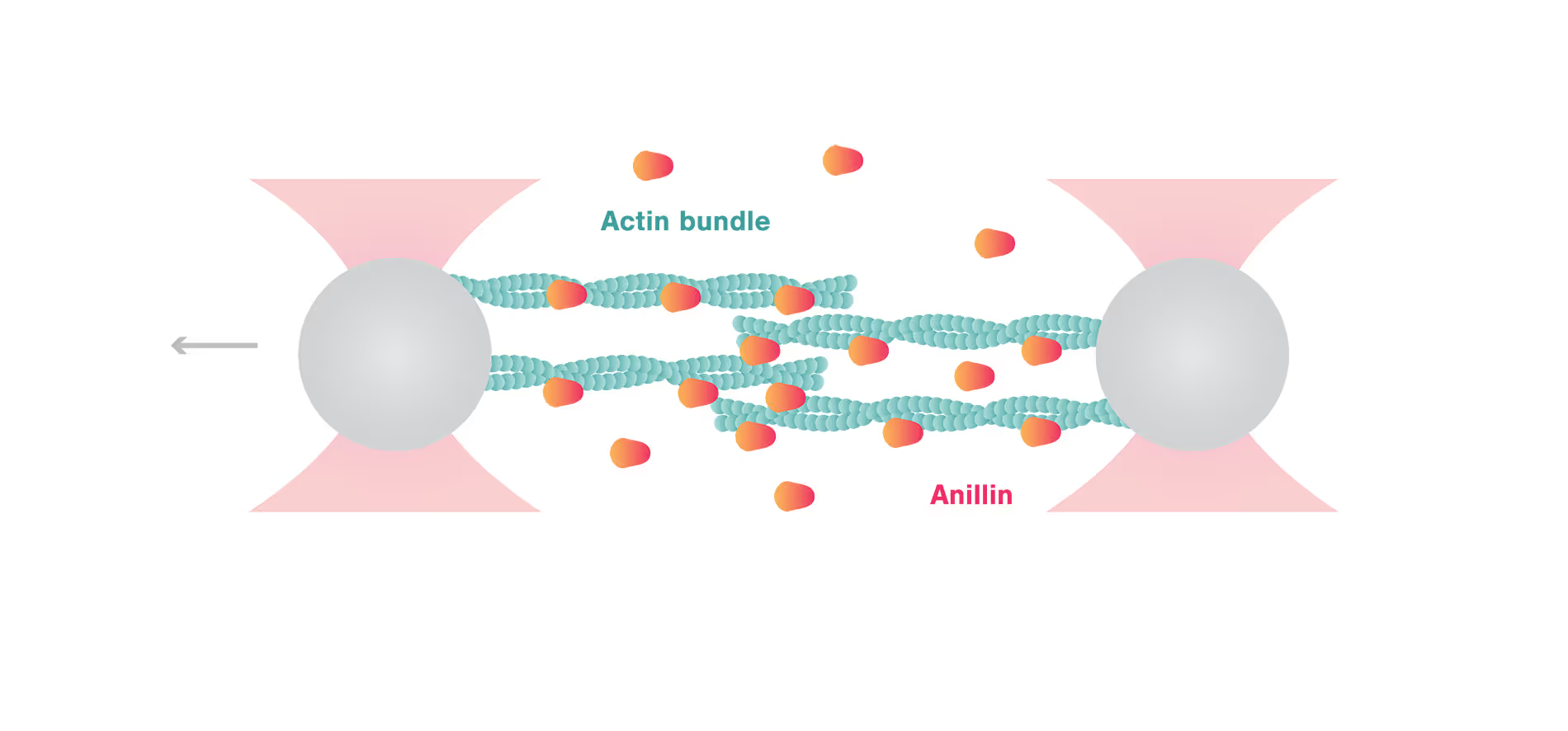
C-Trap uncovers autonomous force generation of anillin contracting actin filaments
Missense mutation in kinesin motor leads to faster detachment from microtubules measured by C-Trap®
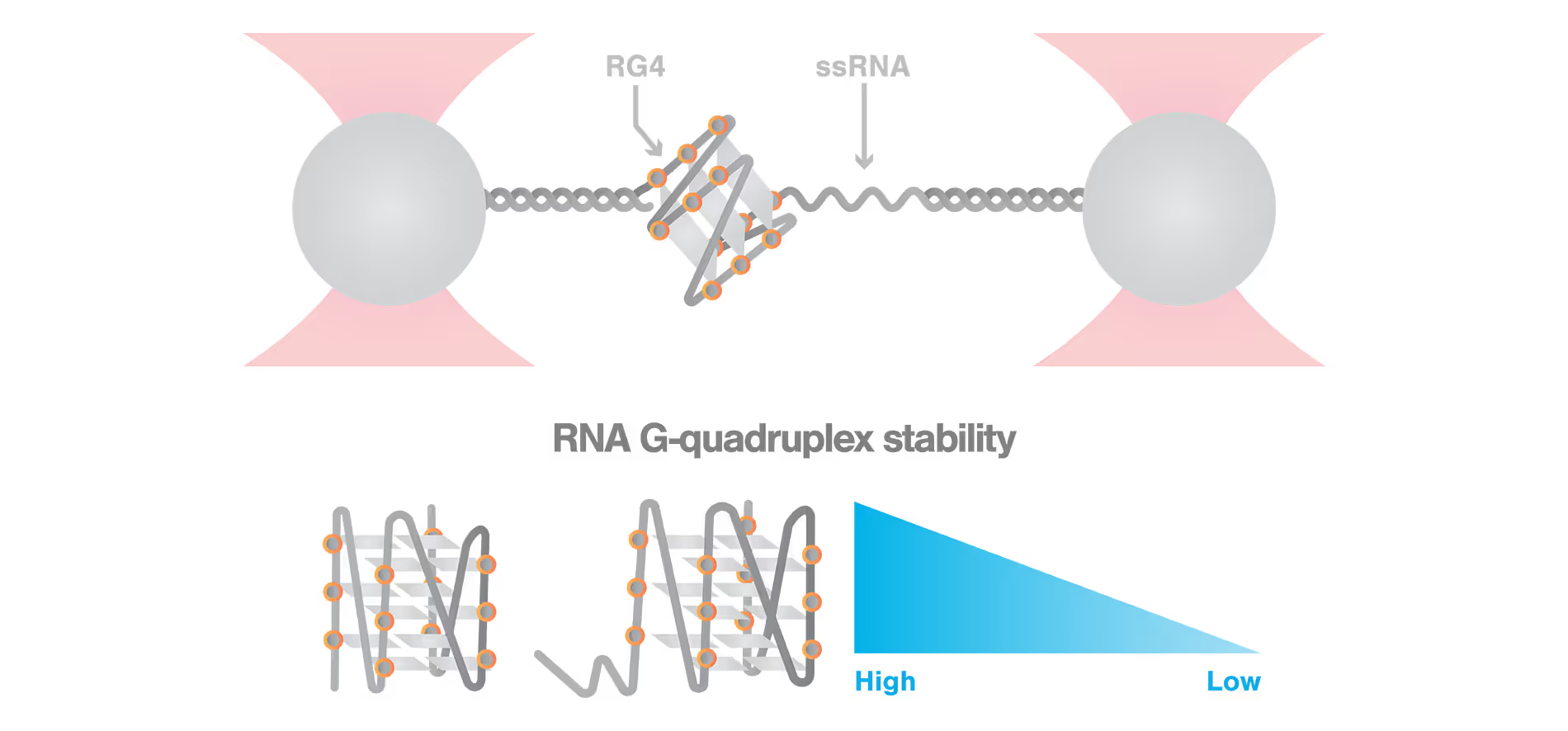
ssRNA significantly decreases RNA G-quadruplex stability measured by C-Trap®
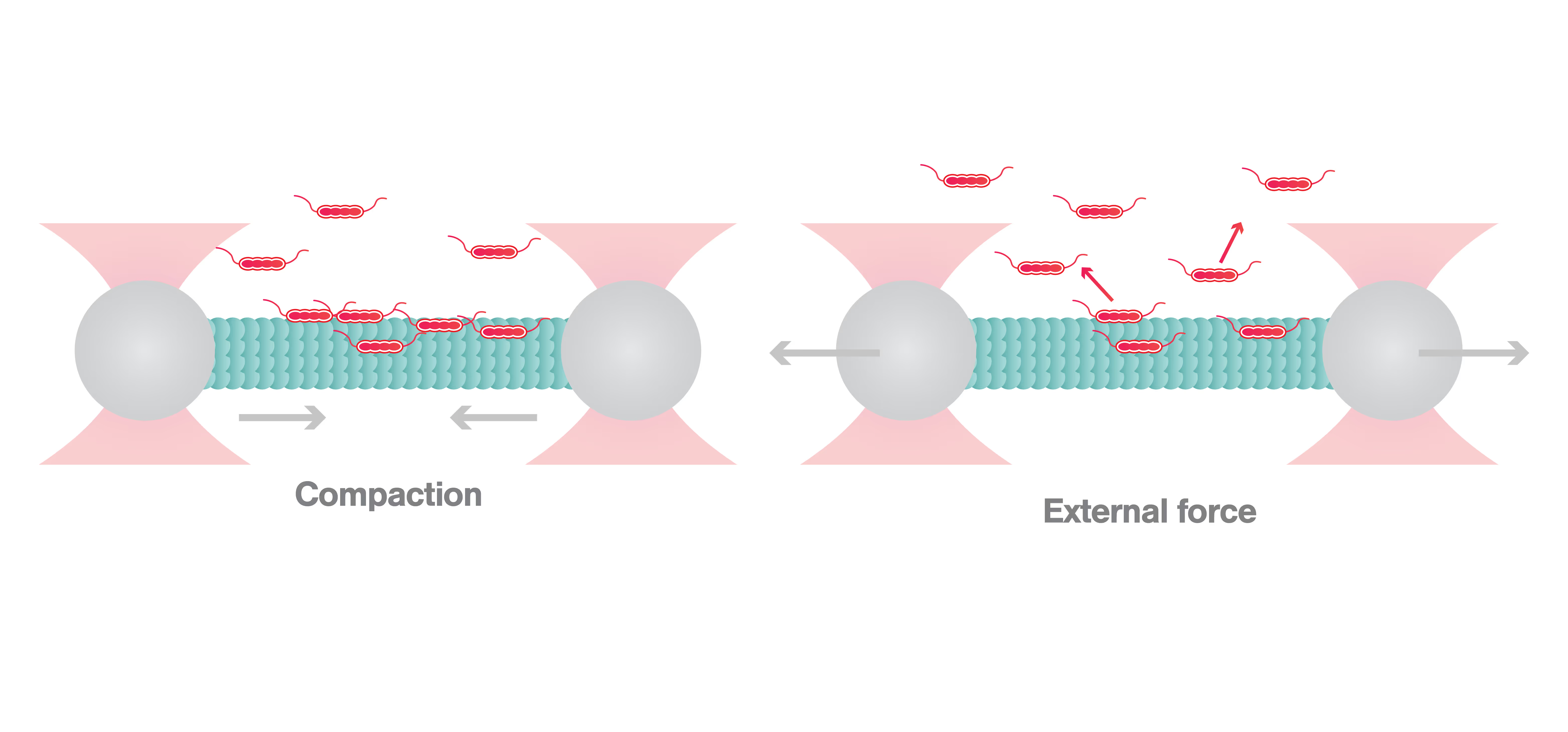
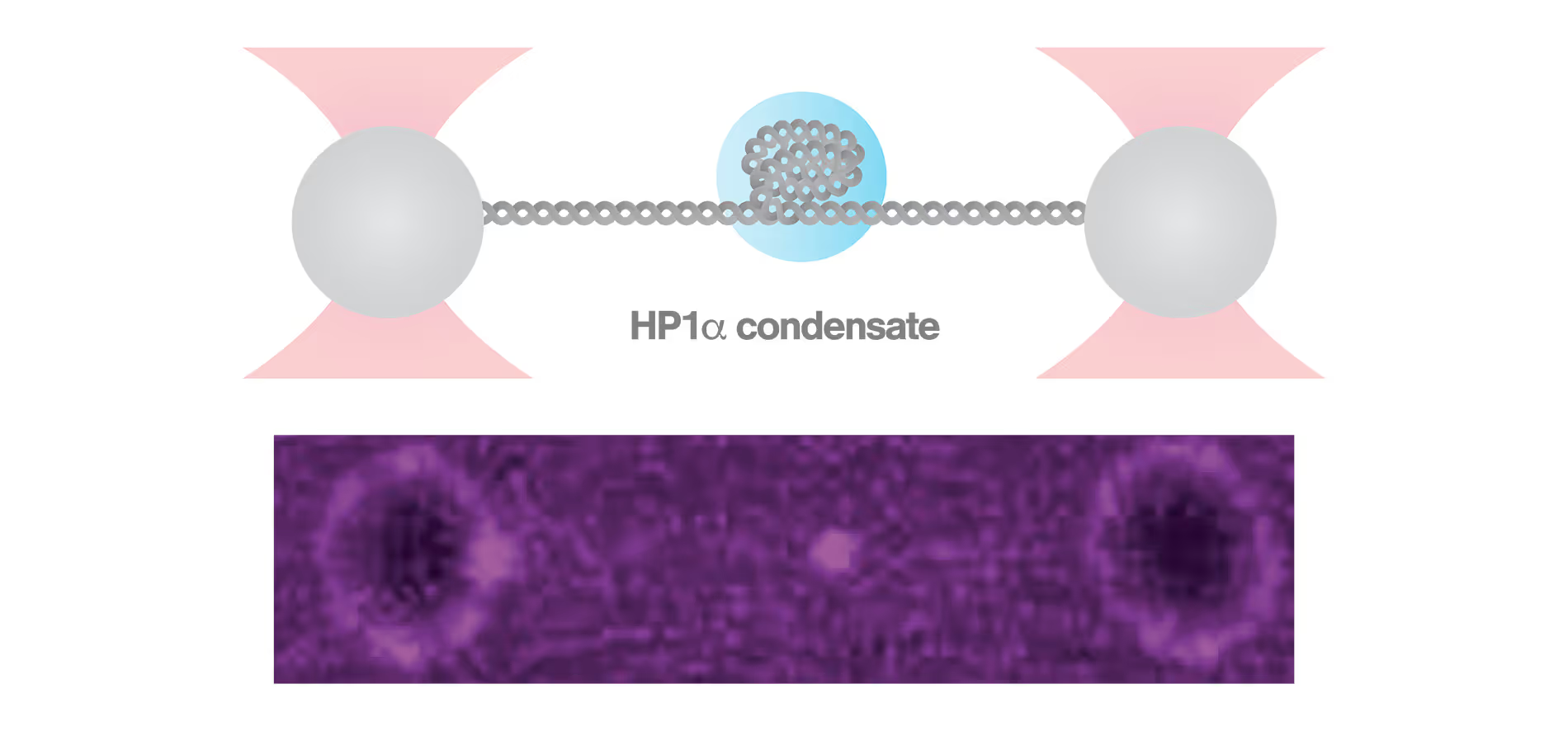
C-Trap reveals how HP1α-mediated heterochromatin modulates its stability in response to applied forces
New insights into the dynamics of initial replisome assembly uncovered by the C-Trap®
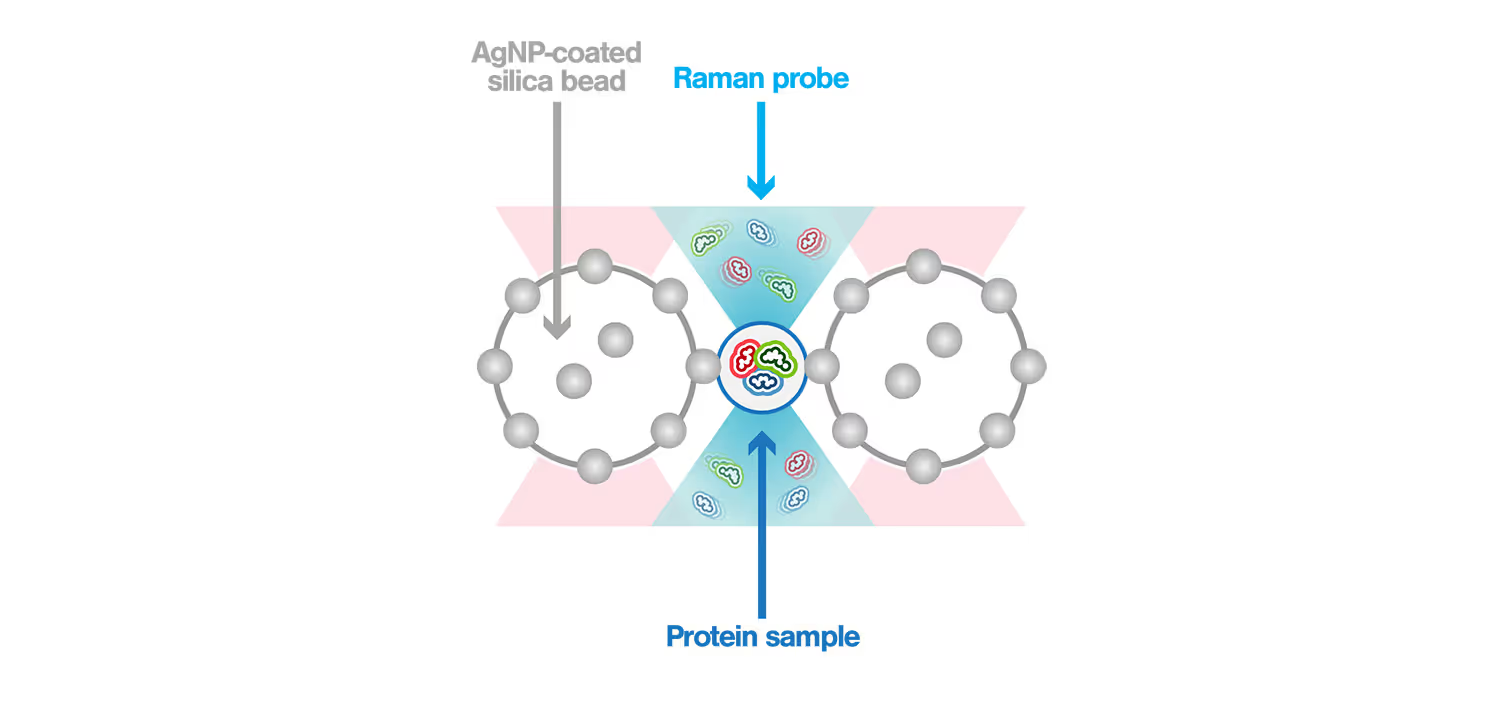
Highly reproducible single-molecule analysis of transient structures of native proteins using m-Trap®dual optical tweezers combined with Raman spectroscopy
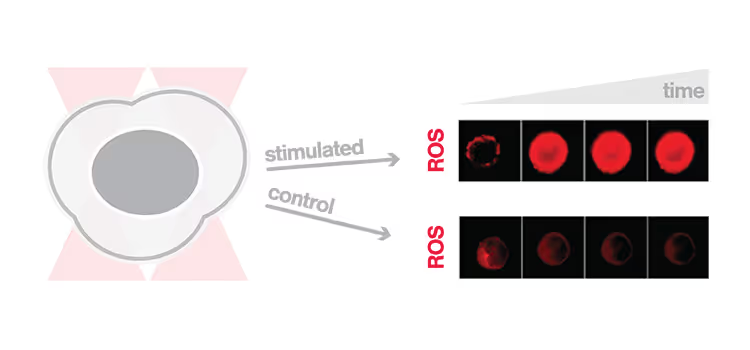
Early activation of single macrophages captured by the C-Trap®
Researchers identify KIF1A properties associated with neuron transport with the C-Trap®
Evaluation of protein droplet viscoelasticity with the C-Trap®reveals new material properties
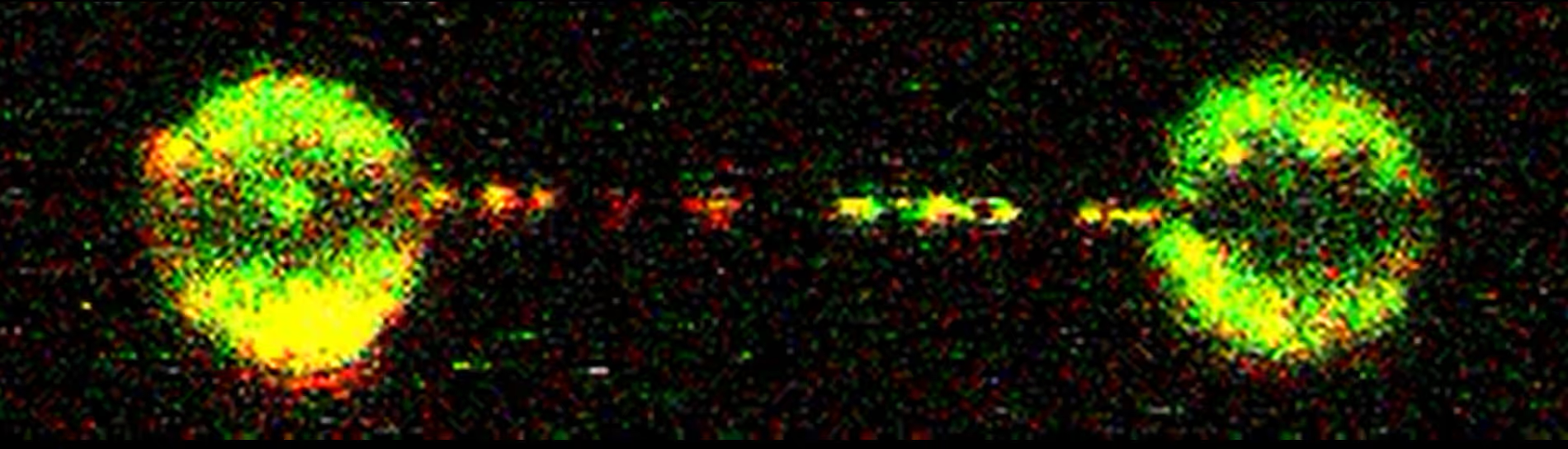
Coordinated stimulation of RAD51 filament assembly and growth revealed with the C-Trap®
Researchers study the viscoelasticity of human cells as a response to external cues using the high-throughput AFS®
The softening effect of vimentin filament phosphorylation measured with the C-Trap®
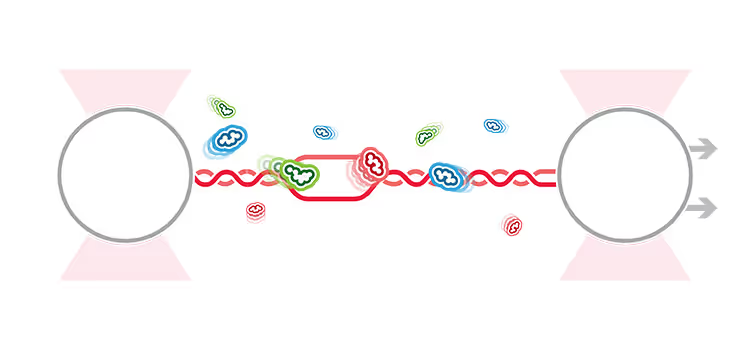
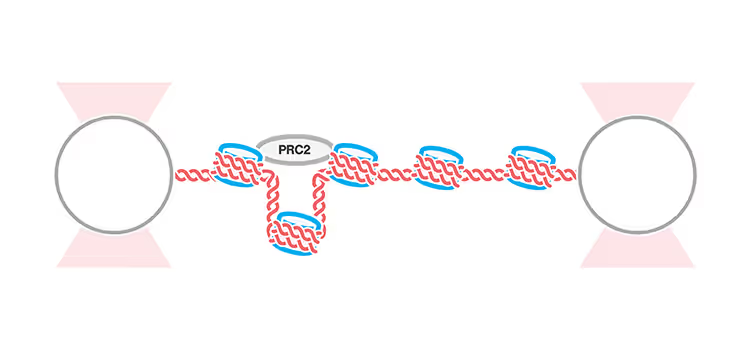
Distal bridging between nucleosomes by polycomb complex PRC2 identified with the C-Trap®
New study highlights interconvertible RecA–ssDNA states with the help of u-Flux™ microfluidics
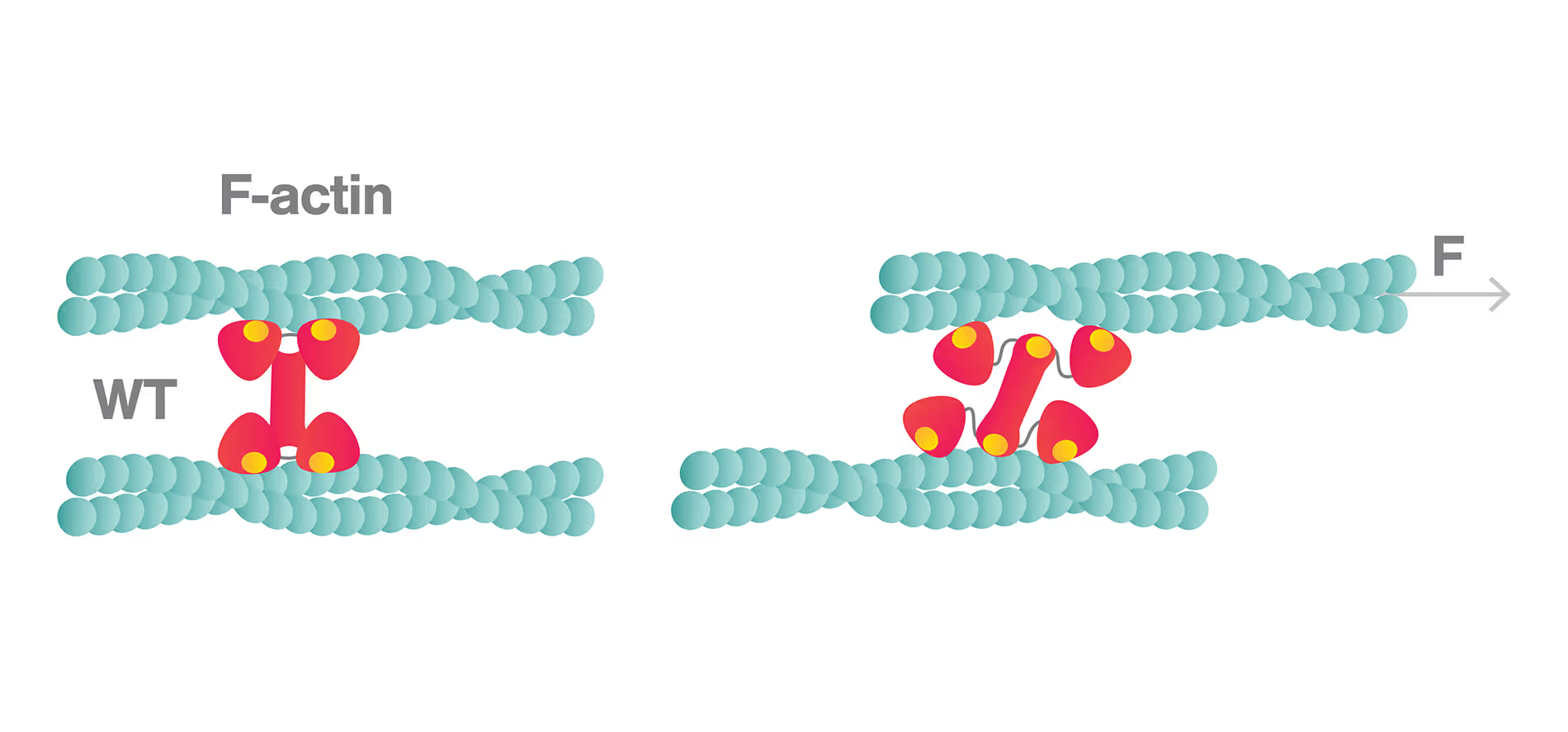
Force-dependent actin filament-binding by α-catenin recorded with the C-Trap®
Researchers use the m-Trap®and C-Trap®to discover CRISPR-Cas9 properties that can help improve in vivo efficiency
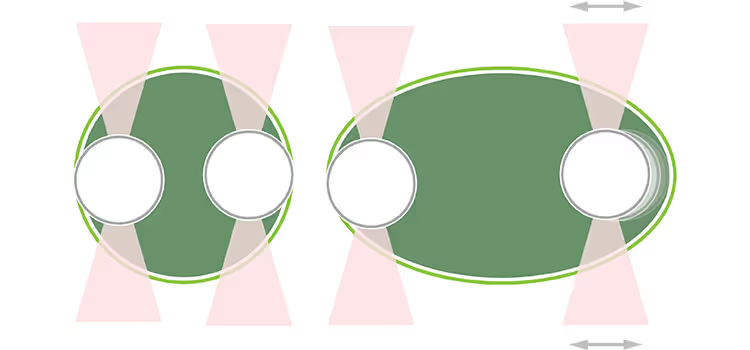
Researchers investigate the effects of biomolecular droplet structure on fusion times using the C-Trap®
Electrostatic effects on vimentin stiffness demonstrated with the C-Trap®
In vitro characterization of nucleosomes upon DNA unwrapping using the C-Trap®
Vesicle-like RNA–protein condensates characterized by the C-Trap®
Exploring mitochondrial translation processes with the C-Trap®
Recent review highlights the impact of force spectroscopy and the C-Trap®
Single-molecule measurements with AFS® and optical tweezers reveal progressive assembly of viruses
New insights into the initiation of nucleotide excision repair found using C-Trap®
New mechanism of helicase-induced DNA unwinding from nicks found with the C-Trap®
Controlled ligand gradient through C-Trap®facilitates DSM measurements
Study shows detailed properties of Ca2+ sensors during membrane fusion
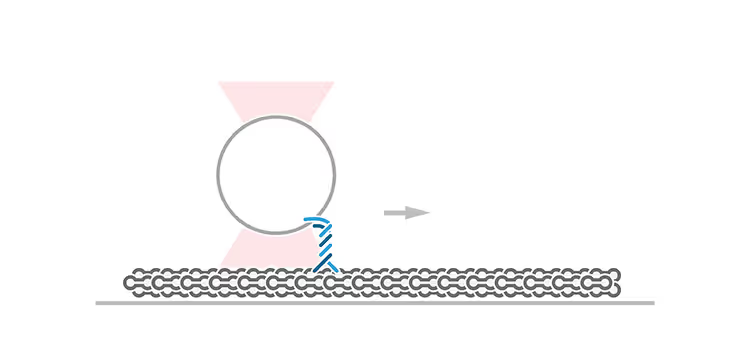
A new DNA–protein coupling procedure improves stability for DSM analyses
Study shows how chaperone enzyme ClpB processively translocates polypeptide loops using C-Trap®correlated optical tweezers and confocal microscopy
Single-molecule study assesses how cohesin complexes bridge DNA molecules using C-Trap®correlated optical tweezers with fluorescence microscopy
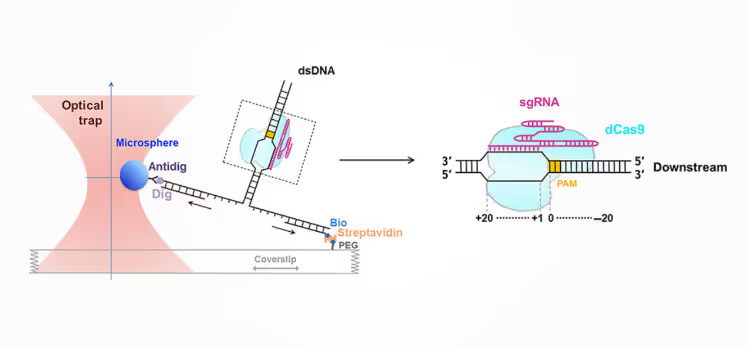
Study finds essential Cas9–DNA interaction downstream of PAM using a single-molecule approach
Study establishes molecular forces responsible for condensation of ribonucleoprotein-RNA complexes
New study maps internal and external structures of red blood cell aggregates by combining AFS® and 3D imaging
New study evaluates the dynamics behind viral assembly at the single-molecule level using both C-Trap™ and AFS®
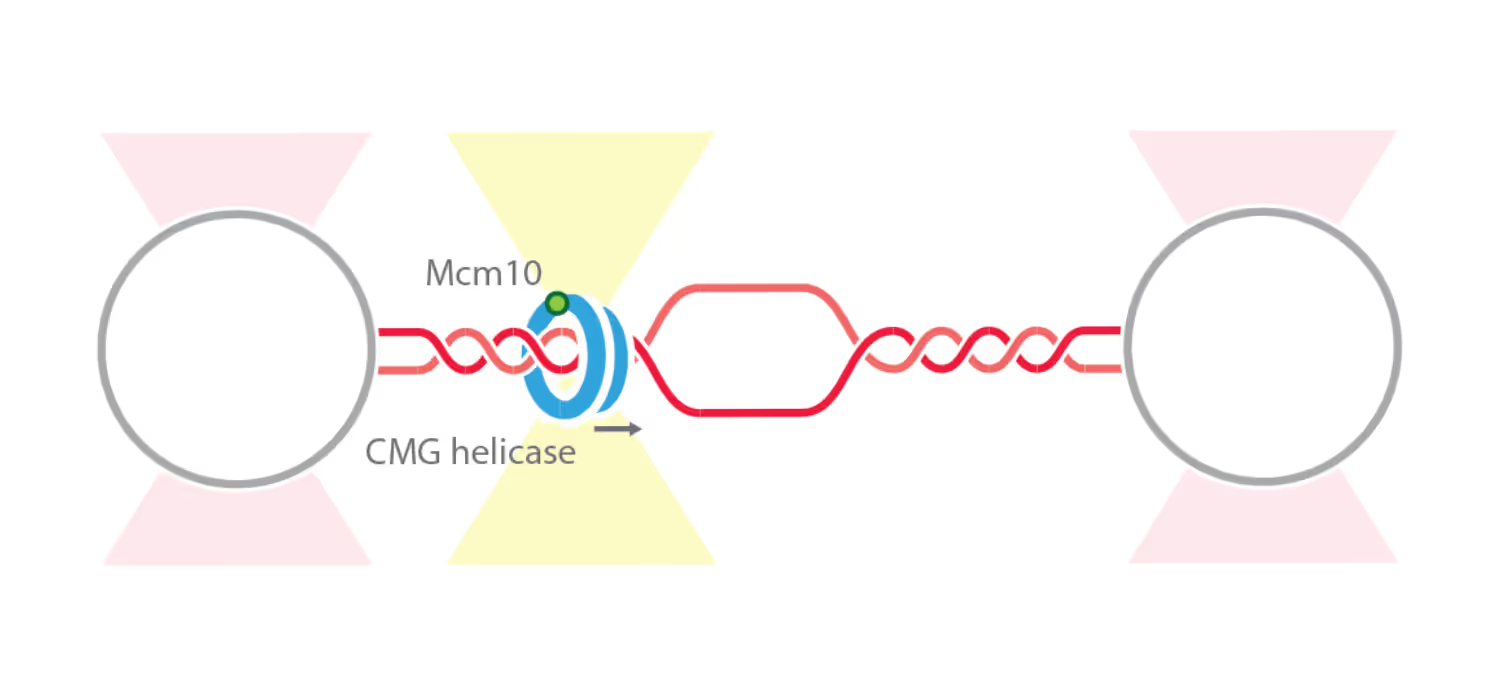
New study resolves helicase dynamics and interactions during DNA replication using C-Trap®
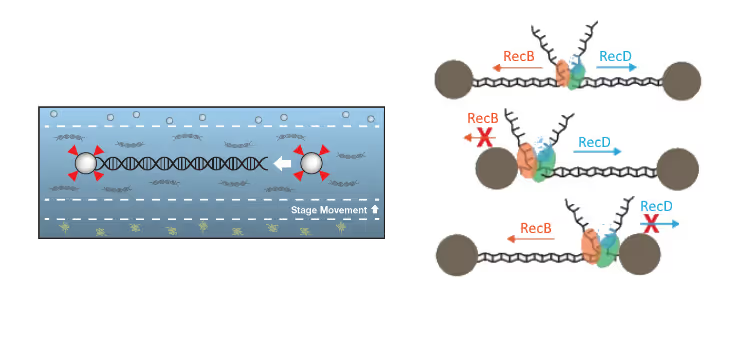
Researchers used u-Flux™ and optical tweezers to measure RecBCD activity
Two papers give new insights on protein droplets using LUMICKS’ C-Trap™
New insights into Cas9 off-target activity
Step into the unresolved with the Surface Assay Toolkit
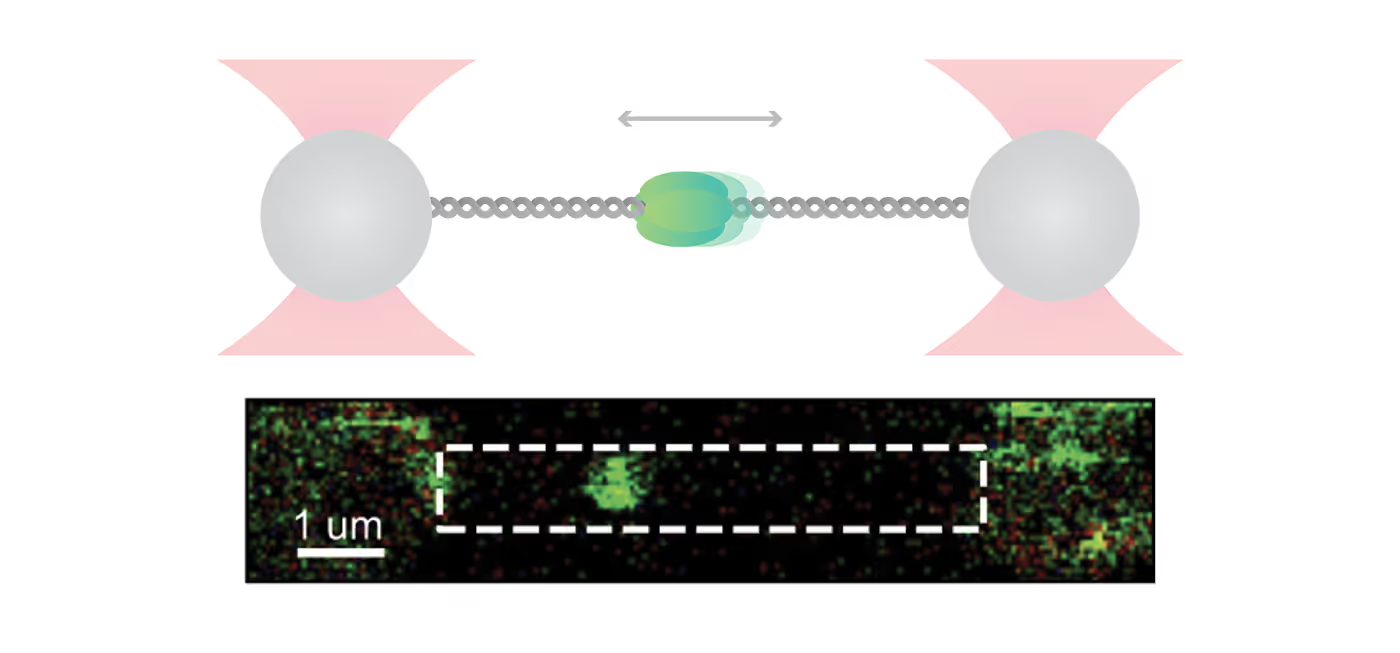
New insights on the formation and dynamics of membraneless organelles using C-Trap™
Deciphering the structure-function relationship of RNA: a complete guide
The 2018 Nobel Prize in Physics
Novel Method for Single-Cell Mechanical Probing using LUMICKS’ AFS™ System
Novel Insights on the Mechanical Properties of IFs using LUMICKS’ Technology
Method to Quantify Local Force Distribution within Biomolecular Systems
Investigation of the Interaction of RAD51 with ssDNA Using CTFM and X-ray Crystallography
Introducing New Generation of Dynamic Single-Molecule Analysis Instruments
Strand-Displacement Polymerization by Reverse Transcriptase Investigated using LUMICKS’ u-Flux™
Filament Manipulation Application Note Now Online
Investigation of the Role of RAD52-DNA Complex in DNA Repair Using Optical Tweezers CTFM and Microfluidics
Study Published on DNA Compaction by Meiotic Protein SYCP3 using LUMICKS Technology
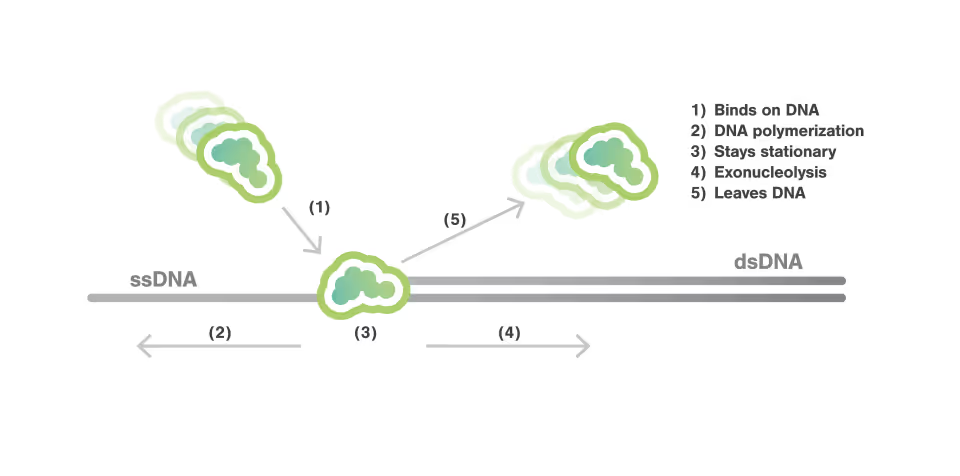
Exonucleolysis and Replication by T7 DNA Polymerase Studied using Optical Tweezers and AFS™
Acinetodin and Klebsidin Lasso Peptides Studied with AFS™ Technology
Effect of Mechanical Stress on Individual Vimentin Filaments Studied using the C-Trap®
Interactions between Single Fibrin Fibers are Measured for the First Time
Paper Using LUMICKS Technology Published in Nature
Study using CTFM Published in Nature Communications
AFS 2.0 Paper Published in Methods
Unparalleled approaches to understand cytoskeletal processes
The cytoskeleton is a critical cell component that regulates cell shape, cell migration, and intracellular organization. It consists primarily of microtubules, intermediate filaments, and actin filaments, each of which play unique roles and contribute to the cytoskeleton’s diverse functionality.
Certain cytoskeletal functions, such as intracellular transport, also require the activity of molecular motors – proteins that catalyze chemical energy into mechanical energy. Alterations in these cytoskeletal components can cause a variety of diseases, ranging from neurological disorders to muscular dystrophies, highlighting a need to better understand the cytoskeleton
Study essential viral processes through a dynamic single-molecule approach
Viral replication strategies and life cycles vary depending on the virus types and can differ throughout all stages, including virus entry, replication, latency, and shedding. Understanding infection and replication processes of viruses is an essential step toward the development of therapeutic strategies to mitigate or treat viral diseases.
Current approaches in virology rely on electron microscopy or RNA and protein quantification to detect viral properties. These methods are either limited to static images, which cannot record dynamic events, or ensemble methods, which are unable to characterize regulatory mechanisms of individual translation or replication processes. Instead, they provide an averaged readout.
In this application note, we show how single-molecule approaches, using optical tweezers correlated with fluorescence and label-free microscopy, can be applied to investigate viral replication processes in detail and aid the development of therapies and virus research as a whole.
Real-time visualization of DNA structural transitions under mechanical stress
Single-molecule force spectroscopy (SMFS) tools are widely used to study structural transitions of DNA during overstretching. However, as many of these tools only provide global information, the exact mechanisms that occur during these transitions remain unclear. Combining SMFS with visualization of local information could resolve this problem. In this application note we will discuss how LUMICKS’ C-Trap® technology combines high-resolution optical tweezers as a SMFS tool with confocal fluorescence microscopy to monitor the transition of double-stranded (ds) DNA to single-stranded (ss) DNA upon applying mechanical stress.
Real-time detection of kinase conformational changes in the presence of a small-molecule inhibitor
This application note introduces you to a new approach for measuring dynamic and highly transient conformational states of proteins and collecting the data in real time. It showcases how to perform these measurements with the C-Trap® Optical Tweezers – Fluorescence and Label-free Microscopy system from start to end. We also introduce you to the adopted features that support and simplify your dynamic single-molecule experiments, regardless of your experience level.
Protein folding and conformational changes
The biological function of macromolecules such as proteins and RNA is intimately linked to their conformation and dynamic structural changes. Proper folding into native conformations is critical for function, while misfolding can lead to loss of activity or the onset of diseases, including neurodegenerative disorders associated with protein aggregation.
Understanding the mechanisms of protein folding and conformational transitions is therefore essential for elucidating biological processes and disease pathways. Single-Molecule Force Spectroscopy (SMFS) offers a powerful approach to probe these mechanisms by isolating individual molecules and monitoring real-time conformational changes under applied mechanical force. This application note presents a demonstrative experiment utilizing high-resolution optical tweezers developed by LUMICKS to investigate the folding pathway of Calmodulin (CaM), the primary calcium-binding protein in the human body.
The results highlight the capability of SMFS to provide high-sensitivity measurements of protein conformational dynamics at the sub-nanometer scale.
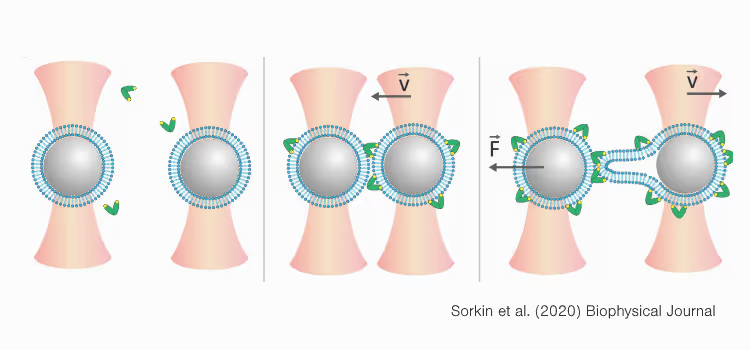
Optical tweezers with fluorescence reveals the dynamics of membrane proteins and droplet fusion
Membrane fusion proteins are critical regulators of cellular membrane dynamics, facilitating essential processes such as intracellular trafficking, exocytosis, and viral entry. To investigate these complex events at the molecular level, correlative force and fluorescence techniques offer unique insights. In this study, we demonstrate the use of LUMICKS’ C-Trap®, which integrates optical tweezers with advanced fluorescence microscopy (confocal, widefield, and STED), to directly quantify and visualize protein-mediated membrane fusion in real time. By coating optically trapped beads with fluorescent lipid bilayers and controlling their proximity, we observe fusion events through both mechanical force changes and correlated fluorescence signals. This dual-modality approach allows high-resolution tracking of membrane stalk formation and fusion kinetics, providing a comprehensive view of the mechanical and structural aspects of membrane fusion. The ability to correlate force measurements with liposomal lumen activity and fluorescence imaging opens new avenues for detailed studies of membrane-protein interactions and fusion dynamics.
Optical tweezers measurements of the DNA conformational dynamics at the single-molecule level
DNA processing reactions such as replication, transcription, and repair rely not only on specific enzymatic interactions but also on the dynamic physical properties of DNA itself. One such dynamic behavior is DNA breathing, or fraying, in which base pairs in double-stranded DNA (dsDNA) spontaneously and transiently separate, forming localized single-stranded DNA (ssDNA) bubbles. These transient bubbles can serve as access points for DNA-binding proteins and are crucial for regulating cellular processes. Due to the fleeting nature of DNA breathing, its study requires highly sensitive tools capable of detecting minute structural changes in real time. In this study, we employed LUMICKS’ high-resolution optical tweezers combined with integrated laminar flow microfluidics to investigate DNA breathing at the single-molecule level. By stretching an 8.4 kbp dsDNA molecule under low-salt conditions and applying a constant tension near the overstretching transition, we observed nanometer-scale force fluctuations indicative of DNA breathing over extended periods. These measurements highlight the capability of LUMICKS’ technology to resolve subtle, dynamic conformational changes in DNA and provide new insights into the physical behavior underlying essential biological processes.
Optical tweezers measurements of protein folding-unfolding and conformational dynamics at the single-protein level
The structure and conformational dynamics of macromolecules such as proteins and RNA are fundamental to their biological function. Proper protein folding into a native state is essential for activity, while misfolding can lead to loss of function or toxic aggregation, a hallmark of many neurodegenerative diseases. To better understand these mechanisms, Single-Molecule Force Spectroscopy (SMFS) offers a powerful approach to directly observe protein folding and conformational transitions in real time. In this application note, we demonstrate the use of LUMICKS’ C-Trap™—a high-resolution optical tweezers and fluorescence microscopy system—to investigate the folding pathway of Calmodulin (CaM), a key calcium-binding protein in humans. By mechanically manipulating individual CaM molecules, we track their conformational changes with sub-nanometer precision, revealing insights into their folding dynamics and the mechanical stability of intermediate states. This study showcases the potential of SMFS as a tool for unraveling complex molecular behaviors at the single-molecule level.
Optical tweezers combined with fluorescence reveals small molecule-protein interactions
Small molecule inhibitors are widely used in the pharmaceutical industry. They are often used in cancer therapy and they still remain one of the most effective agents in clinical use. Intercalation of small molecules within the DNA template, or binding to the active binding sites of various enzymes, are broadly used as drug treatments to compromise DNA associatedprocesses that progress in an abnormal fashion.
By using correlated high-resolution optical tweezers and fluorescence microscopy, not only can the binding properties of small molecules be studied (e.g., kinetics or diffusive features) but also their effect in the inhibition of the activity of DNA processing motors.
Obtain direct evidence of the processes involved in RNA virus replication using a DSM approach
The studies presented here demonstrate the potential of the single-molecule solutions offered by LUMICKS’ C-Trap in the study of RNA virus replication.
Performing single-molecule experiments, the researchers uncovered two opposing and significant roles of RNA binding proteins. Zimmer et al. provided direct
evidence that the RNA-binding protein ZAP-S directly impacts the SARS-CoV-2 1a/1b frameshifting and can thus inhibit viral replication [1] – a discovery that
could lead to the development of novel Coronavirus treatments. Contrarily, Hill et al. showed that accumulation of protein 2A in host cells can lead to increased
viral protein translation through RNA binding [2], allowing for the development of RNA therapies that could help treat encephalomyocarditis.
High throughput DSM measurements of DNA-binding proteins with DNA repeat assembly
Biological mechanisms are not static or homogeneous, which is why dynamic single-molecule analysis has become a powerful tool to gain a more complete understanding of said mechanisms. However, this lack of homogeneity leads to the need to collect a statistically relevant number of data points to gain meaningful insights. The ability to generate high numbers of results was often a shortfall of the first dynamic single-molecule techniques. This is further compounded by traditional DNA-protein interaction studies which often only incorporate single binding sites into the tethered DNA construct. Therefore, observing enough binding events to obtain sufficient statistics to unambiguously prove a molecular mechanism can be time-consuming.
As a leader in dynamic single-molecule analysis technology, LUMICKS has already launched the C-Trap® Optical Tweezers – fluorescence microscopy system, which combines user-friendly sample handling and automated data analysis, greatly accelerating the collection and analysis of data. Further developments in biochemistry by LUMICKS has led to the development of a new DNA repeat assembly kit. This new kit will deliver the next increase in throughput, leading to the generation of statistically relevant results, in a fraction of the previous time required!
Deciphering the structure–function relationship of RNA - a complete guide
With this guide we aim to equip scientists with the necessary knowledge to perform optical tweezers experiments to study the dynamics, structure, and function relationship of RNA molecules.
Analyze Cas9 binding and cleavage properties in real-time while manipulating DNA structures
This application note introduces you to a new dynamic single-molecule analysis approach that enables you to study the binding and cleaving properties of Cas-related complexes and other gene-editing tools. We show results from a recent publication from the laboratory of Prof. David Rueda at the Imperial College in London, UK, which were obtained using the C-Trap®.
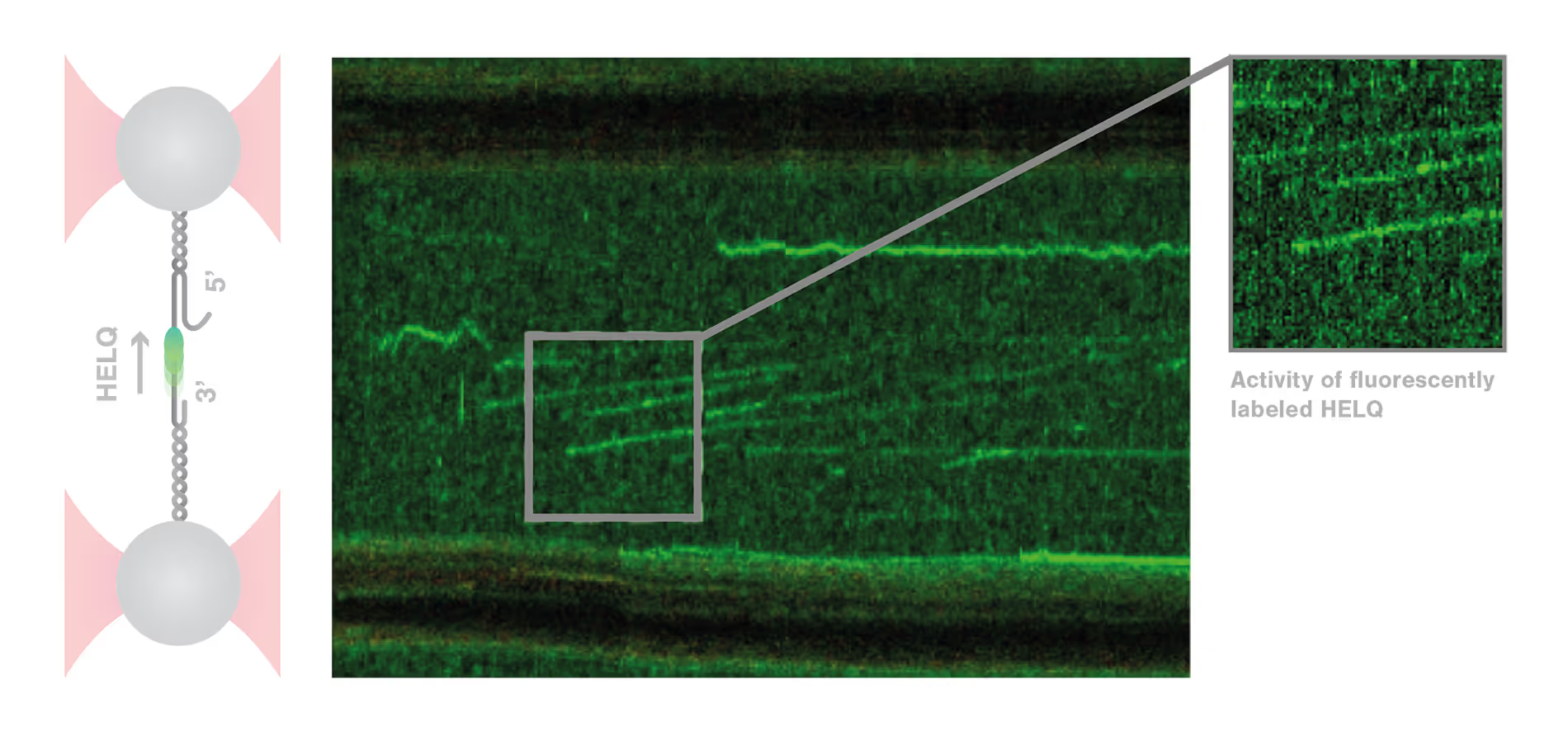
Dynamic single-molecule analysis offers invaluable insights into DNA repair mechanisms: HELQ in DSB case study
DNA repair is a highly complex and dynamic process that involves the interplay of numerous different proteins and components. The helicase HELQ is known to play a role in double-stranded breaks (DSBs) repair, but its molecular mechanisms remain unknown.
A study by the research group led by Simon Boulton presented how dynamic single molecule analysis leads to direct visualization of the mechanism of HELQ in DNA double-stranded breaks (DSBs) repair. Check out this application note to learn more.
Manipulate and study protein droplet dynamic and properties in realtime to understand phase separation
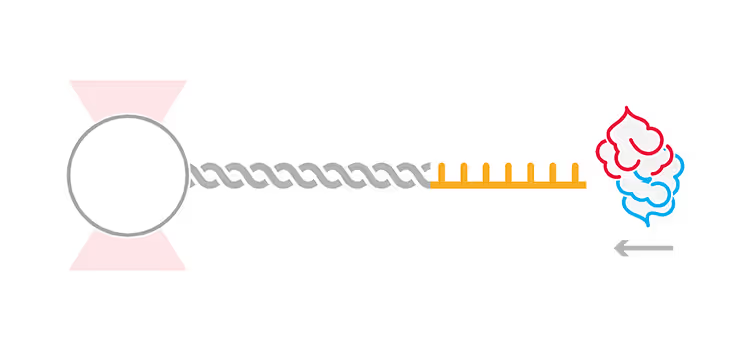
In this application note, we highlight multiple experiments conducted at Banerjee Lab at the University at Buffalo, and Hyman Lab at the Max Planck Institute of Molecular Cell Biology and Genetics on phase separation using the C-Trap dynamic single molecule technology. We present the two approaches the studies used to assess specific properties of protein droplets through applied forces.
Single-molecule visualisation of DNA repair mechanisms and non-homologous end joining (NHEJ)
DNA repair, the collection of highly regulated mechanisms by which a cell identifies and repairs DNA damage, remains one of the most essential processes of human life. Impaired DNA repair systems may lead to malignant mutations that jeopardize cellular well-being.
To study DNA repair, single-molecule studies have proven to
greatly enhance understanding at the molecular level. LUMICKS offers dynamic single-molecule analysis, a novel tool to identify which protein complexes are involved in the non-homologous end joining (NHEJ) DNA repair pathway and their mechanisms.
Unlock the true potential of CRISPR Cas9 technology through real-time direct visualization of Cas9 gene-editing
CRISPR Cas9 is a gene editing tool that has increased in popularity due to its simplicity to use. It allows researchers to seamlessly edit DNA sequences by combining a sequence identifying guide RNA (gRNA) with the Cas9 endonuclease enzyme. However, its applicability as a gene-editing and therapeutic tool is impeded due to undesired off-target binding of Cas9.
Conventional bulk in vitro assays provide information on the DNA sequences but do not give insights into the changes in the local DNA structure. In this application note, dynamic single molecule (DSM) analysis is shown to be a novel and a more powerful investigative tool to directly visualize and manipulate CRISPR Cas9 – DNA interactions, and accelerate gene-editing researchand therapy development.
Golden Gate meets C-Trap: A powerful combination for unprecedented molecular insights
Precisely manipulating genetic material at the single molecule level is gaining importance across life sciences – and so do the tools that allow researchers to do exactly that. The C-Trap system combines single molecule fluorescence microscopy with optical tweezers to manipulate DNA, allowing researchers to directly observe and track molecular events as they occur. Designing and creating specific DNA constructs is crucial for maximizing the potential of single molecule studies. In this application note we introduce the powerful combination of cutting edge biochemistry and single-molecule visualization methods to increase throughput and maximize the results gained from each individual measurement.
Accelerating DNA-protein interaction experiments with nuclear extracts in the C-Trap
Dive into the cutting-edge fusion of nuclear extracts and dynamic single-molecule (DSM) analysis in LUMICKS’ C-Trap. This approach revolutionizes how DNA-protein interactions are studied, offering unmatched biological relevance and accessibility. Biology researchers eager to unravel the complexities of nuclear processes will find this application note a game-changer. Explore how the C-Trap elevates molecular biology research to unprecedented heights.
C-Trap Product Brochure





.avif)