Researchers led by Yael David and Shixin Liu have published a Nature Communications study describing for the first time the mechanisms underlying the co-condensation between transcription factors and DNA. By performing dynamic single-molecule experiments using LUMICKS’ C-Trap® technology, they were able to prove the physiological importance of phase-separated sub-compartments (co-condensates) for gene expression. “This represents an additional, but not mutually exclusive, mechanism for gene regulation besides the canonical sequence-specific transcription factor-DNA interaction paradigm,” according to the authors.
There is an increasing body of evidence pointing to the importance of biomolecular condensates for the control of DNA transcription. However, whether the force exerted by these condensates on DNA is strong enough to be physiologically relevant remains unclear. In this publication, researchers used Sox2—a crucial transcription factor involved in embryogenesis and pluripotency that belongs to the HMG (High Motility Group) superfamily of proteins—that structurally changes DNA by binding to it, dictating cell fate. Sox2 has a globular DNA-binding HMGB domain flanked by IDRs at the C- and N-terminals.
Tuan Nguyen and colleagues showed that Sox2 co-condensates with DNA, sequestering it from the environment, and that these condensates exert mechanical forces driving genome rearrangement. The scientists discovered that the Sox2 HMGB domain drives the condensation, while the IDR domains are crucial for applying force to the DNA.
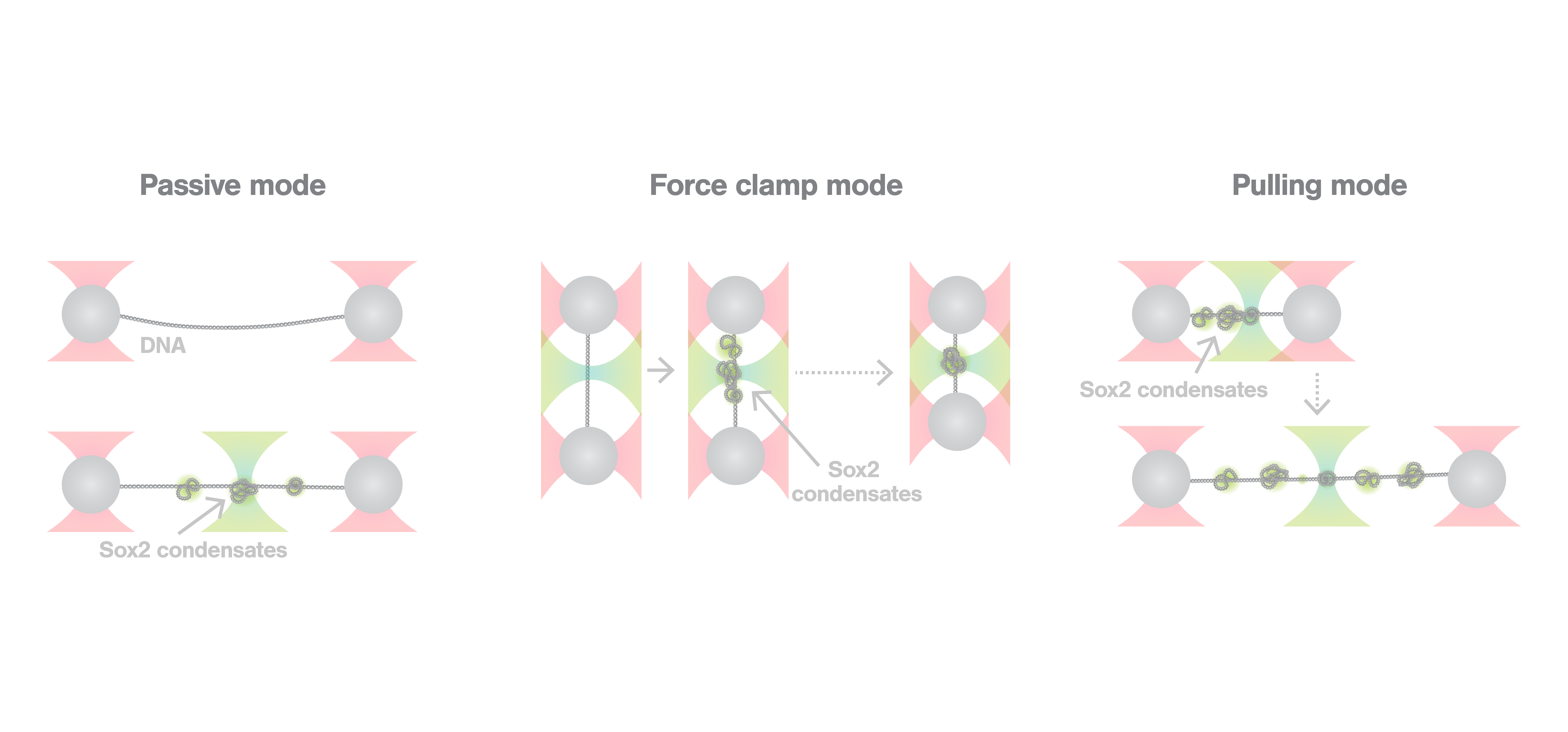
To describe the co-condensation mechanism at a single-molecule level, the researchers went on to combine optical tweezers and scanning confocal microscopy using the C-Trap in 3 different modes: passive, force clamp, and pulling mode. Sox2 was fluorescently labeled to enable the visualization of co-condensate formation in real time, and DNA was tethered between two optically trapped beads. Passive mode experiments revealed that Sox2 condensates form on the DNA, and they exert force on the filaments up to 7pN. Force clamp experiments—where the force between the trapped beads was kept constant keeping the DNA at a constant tension—showed that, when a force as high as 10pN is applied, no condensates are formed. Finally, pulling mode experiments highlighted that, once formed, co-condensates are very stable, as a significant fraction of them remained, even when the DNA was overstretched (65pN). C-Trap experiments enabled researchers to measure and visualize in real time a recently discovered process, co-condensation of transcription factors and DNA, demonstrating its physiological relevance for gene regulation.
As the authors state: “We stress that the force values obtained from the optical tweezers assay represent a more direct and accurate measure of the mechanical tension that Sox2 condensates exert on DNA.”
For further information about this study, check out the paper “Chromatin sequesters pioneer transcription factor Sox2 from exerting force on DNA,” published in Nature Communications.
Are you interested in using dynamic single-molecule tools like the C-Trap for your research? Do not hesitate to contact us for more information, a demo, or a quote.
Top image adapted from Nguyen et al., Nature Communications, 2022.