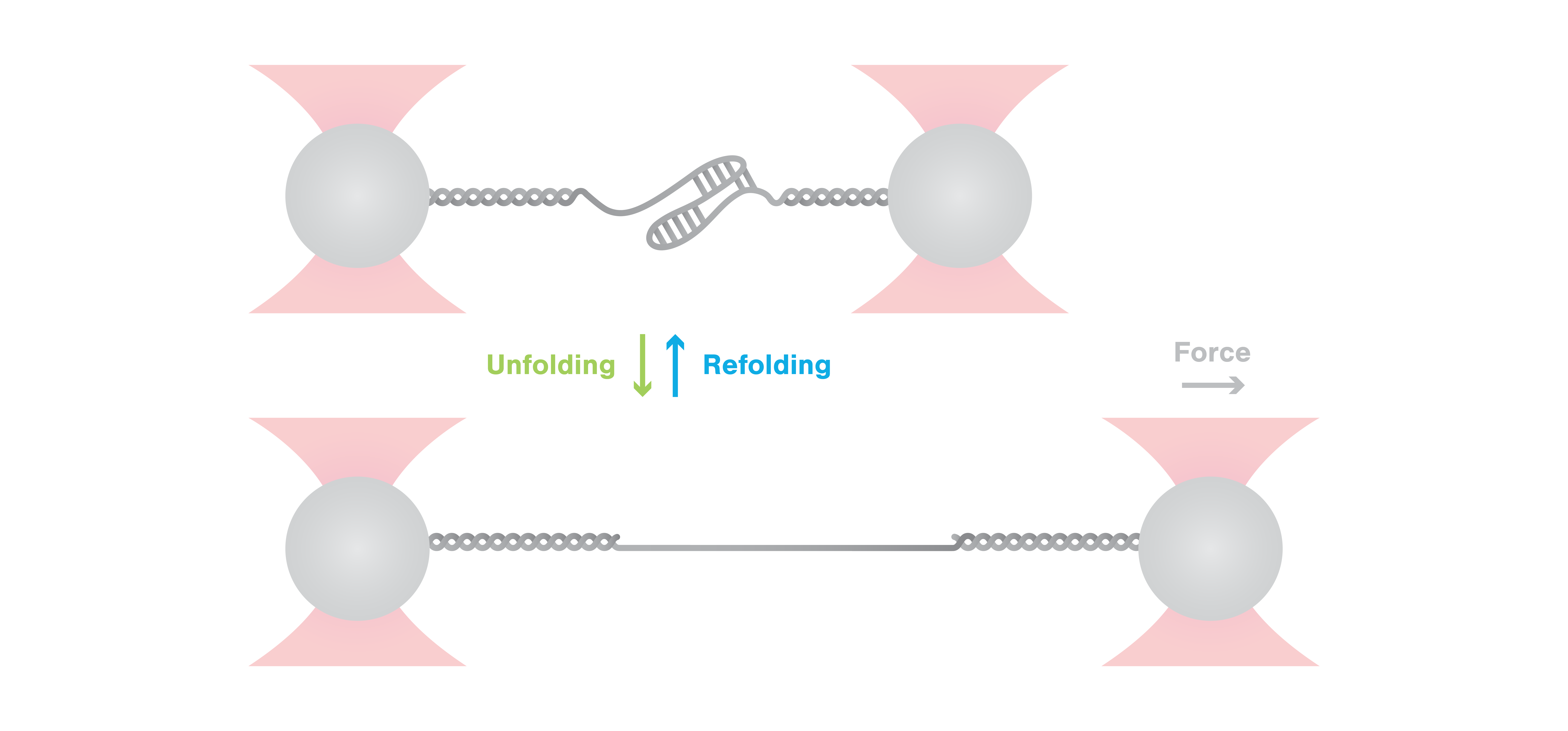
Researchers from the Helmholtz Institute for RNA-based Infection Research (HIRI) in Würzburg, led by Professor Neva Caliskan, recently published a study in Nature Communications, shedding light on the mechanisms that govern programmed ribosomal frameshifting (PRF) in SARS-CoV-2 virus and the role played by host proteins in regulating this process, as demonstrated by C-Trap experiments.
PRF is a mechanism of gene expression used by many viruses to increase their coding potential by shifting the reading frame. Specifically, in SARS-CoV-2 the –1 programmed ribosomal frameshifting (–1PRF) event allows the translation of different proteins from the same transcript and is essential for viral replication and transcription. PRF relies on the presence of a slippery heptameric nucleotide sequence followed by an RNA-specific secondary structure (a putative pseudoknot). Although several interactors are known for PRF RNA, how these factors alter the mechanical properties of RNA and affect the choice of the reading frame remains unclear.
In this study, Matthias Zimmer and coworkers found that the endogenous protein ZAP-S is a direct inhibitor of PRF by SARS-CoV-2 and plays a crucial role in the virulence of this pathogen. A multidisciplinary approach was used to identify the short isoform of zinc-finger antiviral protein (ZAP-S) as a host-encoded inhibitor of SARS-CoV-2 1a/1b frameshifting in vitro and in vivo. Following these findings, the authors hypothesized that ZAP-S might interfere with PRF by targeting the putative pseudoknot in the RNA frameshifting element.
To test this hypothesis and clarify the effect of ZAP-S binding on the structure and mechanical stability of the RNA, the scientists employed single-molecule pulling experiments using the C-Trap. Hereby, they stretched the complex to induce unfolding and relaxed it again to allow refolding of the structure. Strikingly, these experiments showed that ZAP-S did not affect the unfolding of RNA but did prevent refolding of the RNA structure into its native state.
The results presented in this article are of great impact as they provide the first evidence that the host-encoded RNA-binding protein ZAP-S directly impacts the SARS-CoV-2 1a/1b frameshifting and thus can inhibit viral replication. C-Trap single-molecule pulling experiments were hereby deciding in revealing the mechanism of this interference. Next to expanding our understanding of RNA-based regulation mechanisms in viruses, these findings prove ZAP-S to be a suitable target for the development of drugs against SARS-CoV-2 and other viral infections.
To learn the details of this study and find out how ZAP-S interacts with translating ribosomes, read the full article “The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting”, published in Nature Communications.